Lab 5 - Julia Code#
Authors: Valerie Dube, Erzo Garay, Juan Marcos Guerrero y Matias Villalba
# Libraries
using DataFrames
using CSV
using StatsPlots
using Statistics
using StatsModels
using GLM
# Import data and see first observations
df = CSV.read("../../data/processed_esti.csv", DataFrame)
first(df, 5)
| Row | y | w | gender_female | gender_male | gender_transgender | ethnicgrp_asian | ethnicgrp_black | ethnicgrp_mixed_multiple | ethnicgrp_other | ethnicgrp_white | partners1 | postlaunch | msm | age | imd_decile |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Int64 | Int64 | Int64 | Int64 | Int64 | Int64 | Int64 | Int64 | Int64 | Int64 | Int64 | Int64 | Int64 | Int64 | Int64 | |
| 1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 27 | 5 |
| 2 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 19 | 6 |
| 3 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 26 | 4 |
| 4 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 20 | 2 |
| 5 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 24 | 3 |
describe(df)
| Row | variable | mean | min | median | max | nmissing | eltype |
|---|---|---|---|---|---|---|---|
| Symbol | Float64 | Int64 | Float64 | Int64 | Int64 | DataType | |
| 1 | y | 0.351926 | 0 | 0.0 | 1 | 0 | Int64 |
| 2 | w | 0.529615 | 0 | 1.0 | 1 | 0 | Int64 |
| 3 | gender_female | 0.584244 | 0 | 1.0 | 1 | 0 | Int64 |
| 4 | gender_male | 0.413456 | 0 | 0.0 | 1 | 0 | Int64 |
| 5 | gender_transgender | 0.00230017 | 0 | 0.0 | 1 | 0 | Int64 |
| 6 | ethnicgrp_asian | 0.0638298 | 0 | 0.0 | 1 | 0 | Int64 |
| 7 | ethnicgrp_black | 0.0862565 | 0 | 0.0 | 1 | 0 | Int64 |
| 8 | ethnicgrp_mixed_multiple | 0.0885566 | 0 | 0.0 | 1 | 0 | Int64 |
| 9 | ethnicgrp_other | 0.013226 | 0 | 0.0 | 1 | 0 | Int64 |
| 10 | ethnicgrp_white | 0.748131 | 0 | 1.0 | 1 | 0 | Int64 |
| 11 | partners1 | 0.296722 | 0 | 0.0 | 1 | 0 | Int64 |
| 12 | postlaunch | 0.516964 | 0 | 1.0 | 1 | 0 | Int64 |
| 13 | msm | 0.130535 | 0 | 0.0 | 1 | 0 | Int64 |
| 14 | age | 23.1064 | 16 | 23.0 | 30 | 0 | Int64 |
| 15 | imd_decile | 3.47154 | 1 | 3.0 | 9 | 0 | Int64 |
control = select(filter(row -> row[:w] == 0, df), Not(:y))
treatment = select(filter(row -> row[:w] == 1, df), Not(:y))
| Row | w | gender_female | gender_male | gender_transgender | ethnicgrp_asian | ethnicgrp_black | ethnicgrp_mixed_multiple | ethnicgrp_other | ethnicgrp_white | partners1 | postlaunch | msm | age | imd_decile |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Int64 | Int64 | Int64 | Int64 | Int64 | Int64 | Int64 | Int64 | Int64 | Int64 | Int64 | Int64 | Int64 | Int64 | |
| 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 27 | 5 |
| 2 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 26 | 4 |
| 3 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 24 | 3 |
| 4 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 24 | 2 |
| 5 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 24 | 4 |
| 6 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 27 | 2 |
| 7 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 21 | 6 |
| 8 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 18 | 4 |
| 9 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 26 | 2 |
| 10 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 23 | 6 |
| 11 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 24 | 4 |
| 12 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 26 | 3 |
| 13 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 26 | 3 |
| ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ |
| 910 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 28 | 4 |
| 911 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 25 | 2 |
| 912 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 18 | 2 |
| 913 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 26 | 4 |
| 914 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 21 | 3 |
| 915 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 19 | 4 |
| 916 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 21 | 2 |
| 917 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 26 | 3 |
| 918 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 29 | 1 |
| 919 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 27 | 4 |
| 920 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 25 | 4 |
| 921 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 25 | 4 |
using DataFrames, Statistics
# Function to get descriptive statistics
function get_descriptive_stats(group::DataFrame, column::Symbol)
if column == :age
count_val = count(!ismissing, group[:, column])
else
count_val = sum(group[:, column] .== 1)
end
mean_val = mean(group[:, column])
std_val = std(group[:, column])
return count_val, mean_val, std_val
end
get_descriptive_stats (generic function with 1 method)
variables = setdiff(Symbol.(names(df)), [:w, :y])
control_stats = Dict(var => get_descriptive_stats(control, var) for var in variables)
treatment_stats = Dict(var => get_descriptive_stats(treatment, var) for var in variables)
# Convert the dictionary to a DataFrame
control_df = DataFrame(
Variable = collect(keys(control_stats)),
Count = [val[1] for val in values(control_stats)],
Mean = [val[2] for val in values(control_stats)],
Std = [val[3] for val in values(control_stats)]
)
treatment_df = DataFrame(
Variable = collect(keys(treatment_stats)),
Count = [val[1] for val in values(treatment_stats)],
Mean = [val[2] for val in values(treatment_stats)],
Std = [val[3] for val in values(treatment_stats)]
)
control_df = sort!(control_df, :Variable)
treatment_df = sort!(treatment_df, :Variable)
| Row | Variable | Count | Mean | Std |
|---|---|---|---|---|
| Symbol | Int64 | Float64 | Float64 | |
| 1 | age | 921 | 23.1585 | 3.53874 |
| 2 | ethnicgrp_asian | 66 | 0.0716612 | 0.258066 |
| 3 | ethnicgrp_black | 74 | 0.0803474 | 0.271978 |
| 4 | ethnicgrp_mixed_multiple | 78 | 0.0846906 | 0.278572 |
| 5 | ethnicgrp_other | 9 | 0.00977199 | 0.0984226 |
| 6 | ethnicgrp_white | 694 | 0.753529 | 0.43119 |
| 7 | gender_female | 541 | 0.587405 | 0.492569 |
| 8 | gender_male | 377 | 0.409338 | 0.491979 |
| 9 | gender_transgender | 3 | 0.00325733 | 0.0570109 |
| 10 | imd_decile | 36 | 3.46037 | 1.46584 |
| 11 | msm | 114 | 0.123779 | 0.329508 |
| 12 | partners1 | 277 | 0.30076 | 0.458838 |
| 13 | postlaunch | 512 | 0.555917 | 0.497133 |
# Combine control_df and treatment_df into a single DataFrame
combined_df = DataFrame(
Variable = control_df.Variable,
Control_Count = control_df.Count,
Control_Mean = control_df.Mean,
Control_Std = control_df.Std,
Treatment_Count = treatment_df.Count,
Treatment_Mean = treatment_df.Mean,
Treatment_Std = treatment_df.Std
)
# Round numerical columns to 2 decimal places
function round_df(df::DataFrame, decimals::Int)
rounded_df = copy(df)
for col in names(df)[2:end]
rounded_df[!, col] = round.(df[!, col], digits=decimals)
end
return rounded_df
end
formatted_table = round_df(combined_df, 2)
# Print the formatted table
println("Table 1: Descriptive Statistics and Balance\n")
show(stdout, "text/plain", formatted_table)
Table 1: Descriptive Statistics and Balance
13×7 DataFrame
Row │ Variable Control_Count Control_Mean Control_Std Treatment_Count Treatment_Mean Treatment_Std
│ Symbol Float64 Float64 Float64 Float64 Float64 Float64
─────┼────────────────────────────────────────────────────────────────────────────────────────────────────────────────────
1 │ age 818.0 23.05 3.59 921.0 23.16 3.54
2 │ ethnicgrp_asian 45.0 0.06 0.23 66.0 0.07 0.26
3 │ ethnicgrp_black 76.0 0.09 0.29 74.0 0.08 0.27
4 │ ethnicgrp_mixed_multiple 76.0 0.09 0.29 78.0 0.08 0.28
5 │ ethnicgrp_other 14.0 0.02 0.13 9.0 0.01 0.1
6 │ ethnicgrp_white 607.0 0.74 0.44 694.0 0.75 0.43
7 │ gender_female 475.0 0.58 0.49 541.0 0.59 0.49
8 │ gender_male 342.0 0.42 0.49 377.0 0.41 0.49
9 │ gender_transgender 1.0 0.0 0.03 3.0 0.0 0.06
10 │ imd_decile 37.0 3.48 1.49 36.0 3.46 1.47
11 │ msm 113.0 0.14 0.35 114.0 0.12 0.33
12 │ partners1 239.0 0.29 0.46 277.0 0.3 0.46
13 │ postlaunch 387.0 0.47 0.5 512.0 0.56 0.5
The observations for each variable are generally balanced between the control and treatment groups. Additionally, most participants are white, with an average age of approximately 23. The mean IMD decile scores are around 3.5, indicating that participants in both groups tend to come from more deprived areas.
using DataFrames
using StatsPlots
# Group by 'w' and 'gender_male' and count occurrences
df_grouped = combine(groupby(df, [:w, :gender_male]), nrow => :count)
# Calculate the proportion of each group
df_grouped[!, :prop] .= df_grouped.count ./ sum(df_grouped.count)
df_grouped
| Row | w | gender_male | count | prop |
|---|---|---|---|---|
| Int64 | Int64 | Int64 | Float64 | |
| 1 | 0 | 0 | 476 | 0.273721 |
| 2 | 0 | 1 | 342 | 0.196665 |
| 3 | 1 | 0 | 544 | 0.312823 |
| 4 | 1 | 1 | 377 | 0.216791 |
using DataFrames
using Plots
# Extracting the data
w = df_grouped.w
gender_male = df_grouped.gender_male
prop = df_grouped.prop
# Creating a grouped bar plot
groupedbar(
string.(gender_male),
prop,
group = string.(w),
xlabel = "Gender Male (0 = Female, 1 = Male)",
ylabel = "Proportion",
title = "Grouped Bar Graph of Proportion by w and Gender",
legend = :topright,
bar_width = 0.5,
label = ["Treatment = 0" "Treatment = 1"]
)
# Display the plot
plot!(current())
using DataFrames
using StatsPlots
# Group by 'w' and 'partners1' and count occurrences
df_grouped = combine(groupby(df, [:w, :partners1]), nrow => :count)
# Calculate the proportion of each group
df_grouped[!, :prop] .= df_grouped.count ./ sum(df_grouped.count)
# Extracting the data
w = df_grouped.w
partners1 = df_grouped.partners1
prop = df_grouped.prop
# Creating a grouped bar plot
groupedbar(
string.(partners1),
prop,
group = string.(w),
xlabel = "Number of partners (one partner=1)",
ylabel = "Proportion",
title = "Proportion of Partners by Treatment Group",
legend = :topright,
bar_width = 0.5,
label = ["Treatment = 0" "Treatment = 1"]
)
# Display the plot
plot!(current())
using DataFrames
using StatsPlots
# Create density plots for each group in 'w'
density(
df.age,
group = df.w,
xlabel = "Age",
ylabel = "Percentage",
title = "Density Plot of Age by Treatment Group",
legend = :topright,
normalize = true,
label = ["Control" "Treatment"]
)
# Display the plot
plot!(current())
using DataFrames
using StatsPlots
# Group by 'imd_decile' and 'w' and count occurrences
df_grouped = combine(groupby(df, [:imd_decile, :w]), nrow => :count)
# Calculate the proportion of each group within 'imd_decile'
df_grouped[!, :prop] .= df_grouped.count ./ sum(df_grouped.count) * 100 # Convert to percentage
# Create a density-like bar plot
@df df_grouped groupedbar(
:imd_decile,
:prop,
group = :w,
xlabel = "IMD Decile",
ylabel = "Percentage",
title = "Density Plot of IMD Decile by Treatment Group",
legend = :topright,
bar_width = 0.8,
label = ["Control" "Treatment"]
)
# Display the plot
plot!(current())
using DataFrames
using StatsModels
using GLM
model_1 = @formula(y ~ w)
est_1 = lm(model_1, df)
print(est_1)
StatsModels.TableRegressionModel{LinearModel{GLM.LmResp{Vector{
Float64}}, GLM.DensePredChol{Float64, LinearAlgebra.CholeskyPivoted{Float64, Matrix
{Float64}, Vector{Int64}}}}, Matrix{Float64}}
y ~ 1 + w
Coefficients:
────────────────────────────────────────────────────────────────────────
Coef. Std. Error t Pr(>|t|) Lower 95% Upper 95%
────────────────────────────────────────────────────────────────────────
(Intercept) 0.211491 0.0160531
13.17 <1e-37 0.180006 0.242977
w 0.265164
0.0220586 12.02 <1e-31
0.2219 0.308429
────────────────────────────────────────────────────────────────────────
model_2 = @formula(y ~ w + age + gender_female + ethnicgrp_white + ethnicgrp_black + ethnicgrp_mixed_multiple + partners1 + postlaunch + imd_decile)
est_2 = lm(model_2, df)
print(est_2)
StatsModels.TableRegressionModel{LinearModel{GLM.LmResp{Vector{Float64}},
GLM.DensePredChol{Float64, LinearAlgebra.CholeskyPivoted{Float64, Matrix{Float64}
, Vector{Int64}}}}, Matrix{Float64}}
y ~ 1 + w + age + gender_female + ethnicgrp_white + ethnicgrp_black + ethnicgrp_mixed_multiple + partners1 + postlaunch + imd_decile
Coefficients:
───────────────────────────────────────────────────────────────────────────────────────────
Coef. Std. Error t Pr(>|t|) Lower 95% Upper 95%
───────────────────────────────────────────────────────────────────────────────────────────
(Intercept) -0.163972 0.0880134 -1.86
0.0626 -0.336596 0.00865184
w 0.255827 0.0217719 11.75 <1e-29 0.213125 0.298529
age 0.0124373 0.00316801 3.93 <1e-04 0.0062238 0.0186509
gender_female 0.0928923 0.0223094 4.16 <1e-04 0.0491361 0.136649
ethnicgrp_white 0.0498876 0.0411584 1.21 0.2256 -0.0308379 0.130613
ethnicgrp_black -0.0397451 0.0541118 -0.73 0.4627 -0.145877 0.0663863
ethnicgrp_mixed_multiple -0.035726 0.0534103 -0.67 0.5037 -0.140481 0.0690296
partners1 -0.0590884 0.0242699 -2.43 0.0150 -0.10669 -0.0114868
postlaunch 0.0770255 0.0225539 3.42 0.0007 0.0327897 0.121261
imd_decile -0.00410827 0.0074161 -0.55 0.5797 -0.0186537 0.0104372
───────────────────────────────────────────────────────────────────────────────────────────
3. Non-Linear Methods DML#
using Pkg
Pkg.add("DataFrames")
Pkg.add("CSV")
Pkg.add("Statistics")
Pkg.add("StatsModels")
Pkg.add("GLM")
Pkg.add("MLJ")
Pkg.add("XGBoost")
Pkg.add("MLJXGBoostInterface")
Pkg.add("MLJGLMInterface")
Pkg.add("Lasso")
Pkg.add("GLMNet")
Pkg.add("CovarianceMatrices")
Resolving package versions...
No Changes to `C:\Users\Matias Villalba\.julia\environments\v1.10\Project.toml`
No Changes to `C:\Users\Matias Villalba\.julia\environments\v1.10\Manifest.toml`
Precompiling
project...
using DataFrames
using CSV
using Statistics
using StatsModels
using GLM
using Random
using StatsBase
using LinearAlgebra
Precompiling CSV
✓ CSV
1 dependency successfully precompiled in 9 seconds. 26 already precompiled.
Precompiling StatsModels
? StatsFuns
Info Given StatsModels was explicitly requested, output will be shown live
WARNING: Method definition (::Type{Base.MPFR.BigFloat})(Base.Irrational{:twoπ}) in module IrrationalConstants at irrationals.jl:223 overwritten in module StatsFuns on the same line (check for duplicate calls to `include`).
ERROR: Method overwriting is not permitted during Module precompilation. Use `__precompile__(false)` to opt-out of precompilation.
? StatsModels
[ Info: Precompiling StatsModels [3eaba693-59b7-5ba5-a881-562e759f1c8d]
WARNING: Method definition (::Type{Base.MPFR.BigFloat})(Base.Irrational{:twoπ}) in module IrrationalConstants at irrationals.jl:223 overwritten in module StatsFuns on the same line (check for duplicate calls to `include`).
ERROR: Method overwriting is not permitted during Module precompilation. Use `__precompile__(false)` to opt-out of precompilation.
[ Info: Skipping precompilation since __precompile__(false). Importing StatsModels [3eaba693-59b7-5ba5-a881-562e759f1c8d].
Precompiling StatsFuns
Info Given StatsFuns was explicitly requested, output will be shown live
WARNING: Method definition (::Type{Base.MPFR.BigFloat})(Base.Irrational{:twoπ}) in module IrrationalConstants at irrationals.jl:223 overwritten in module StatsFuns on the same line (check for duplicate calls to `include`).
ERROR: Method overwriting is not permitted during Module precompilation. Use `__precompile__(false)` to opt-out of precompilation.
? StatsFuns
[ Info: Precompiling StatsFuns [4c63d2b9-4356-54db-8cca-17b64c39e42c]
WARNING: Method definition (::Type{Base.MPFR.BigFloat})(Base.Irrational{:twoπ}) in module IrrationalConstants at irrationals.jl:223 overwritten in module StatsFuns on the same line (check for duplicate calls to `include`).
ERROR: Method overwriting is not permitted during Module precompilation. Use `__precompile__(false)` to opt-out of precompilation.
[ Info: Skipping precompilation since __precompile__(false). Importing StatsFuns [4c63d2b9-4356-54db-8cca-17b64c39e42c].
Precompiling GLM
? StatsFuns
? Distributions
? StatsModels
Info Given GLM was explicitly requested, output will be shown live
WARNING: Method definition (::Type{Base.MPFR.BigFloat})(Base.Irrational{:twoπ}) in module IrrationalConstants at irrationals.jl:223 overwritten in module StatsFuns on the same line (check for duplicate calls to `include`).
ERROR: Method overwriting is not permitted during Module precompilation. Use `__precompile__(false)` to opt-out of precompilation.
? GLM
[ Info: Precompiling GLM [38e38edf-8417-5370-95a0-9cbb8c7f171a]
┌ Warning: Module StatsFuns with build ID ffffffff-ffff-ffff-0001-616a4687cd8a is missing from the cache.
│ This may mean StatsFuns [4c63d2b9-4356-54db-8cca-17b64c39e42c] does not support precompilation but is imported by a module that does.
└ @ Base loading.jl:1948
[ Info: Skipping precompilation since __precompile__(false). Importing GLM [38e38edf-8417-5370-95a0-9cbb8c7f171a].
Precompiling Distributions
? StatsFuns
Info Given Distributions was explicitly requested, output will be shown live
WARNING: Method definition (::Type{Base.MPFR.BigFloat})(Base.Irrational{:twoπ}) in module IrrationalConstants at irrationals.jl:223 overwritten in module StatsFuns on the same line (check for duplicate calls to `include`).
ERROR: Method overwriting is not permitted during Module precompilation. Use `__precompile__(false)` to opt-out of precompilation.
? Distributions
[ Info: Precompiling Distributions [31c24e10-a181-5473-b8eb-7969acd0382f]
┌ Warning: Module StatsFuns with build ID ffffffff-ffff-ffff-0001-616a4687cd8a is missing from the cache.
│ This may mean StatsFuns [4c63d2b9-4356-54db-8cca-17b64c39e42c] does not support precompilation but is imported by a module that does.
└ @ Base loading.jl:1948
[ Info: Skipping precompilation since __precompile__(false). Importing Distributions [31c24e10-a181-5473-b8eb-7969acd0382f].
# Load the CSV file into a DataFrame
file_path = "C:\\Users\\juanl\\OneDrive\\Desktop\\hg\\data\\processed_esti.csv"
DML = CSV.read(file_path, DataFrame)
Random.seed!(1234)
# Split the data into training and testing sets
n = nrow(DML)
training = sample(1:n, round(Int, 0.75 * n), replace=false)
data_train = DML[training, :]
data_test = DML[setdiff(1:n, training), :]
# Extract the test outcome
Y_test = data_test.y
# Extract matrices for outcome, treatment, and controls
y = reshape(data_train[:, 1], :, 1) # outcome: growth rate
d = reshape(data_train[:, 2], :, 1) # treatment: initial wealth
x = Matrix(data_train[:, Not([1, 2])]) # controls: country characteristics
# Display the first few rows to verify
println("First few rows of y:")
println(y[1:5, :])
println("First few rows of d:")
println(d[1:5, :])
println("First few rows of x:")
println(x[1:5, :])
First few rows of y:
[0; 0; 0; 0; 1;;]
First few rows of d:
[1; 1; 0; 1; 1;;]
First few rows of x:
[1 0 0 0 0 0 0 1 1 0 0 25 6; 0 1 0 0 0 0 0 1 0 0 1 28 5; 1 0 0 0 0 0 0 1 1 1 0 17 6; 1 0 0 0 0 0 0 1 0 1 0 25 2; 1 0 0 0 0 0 0 1 0 0 0 23 2]
using Pkg
Pkg.add("DecisionTree")
using DecisionTree
Resolving package versions...
Installed ScikitLearnBase ─ v0.5.0
Installed DecisionTree ──── v0.12.4
Updating `C:\Users\juanl\.julia\environments\v1.10\Project.toml`
[7806a523] + DecisionTree v0.12.4
Updating `C:\Users\juanl\.julia\environments\v1.10\Manifest.toml`
[7806a523] + DecisionTree v0.12.4
[6e75b9c4] + ScikitLearnBase v0.5.0
Precompiling project...
? StatsFuns
✓ ScikitLearnBase
✓ ColorVectorSpace → SpecialFunctionsExt
✗ PyCall
✗ BinaryProvider
✓ Latexify → DataFramesExt
✗ MLJXGBoostInterface
✓ DecisionTree
✗ Snappy
? StatsModels
? Distributions
✗ ExcelReaders
✗ Parquet
? GLM
? CovarianceMatrices
✗ ExcelFiles
? GLMNet
✗ ParquetFiles
? MLJGLMInterface
? MLJBase
? Lasso
✗ Queryverse
? MLJIteration
? MLJModels
? MLJTuning
? MLJSerialization
? MLJEnsembles
? MLJ
4 dependencies successfully precompiled in 42 seconds. 284 already precompiled.
12 dependencies precompiled but different versions are currently loaded. Restart julia to access the new versions
15 dependencies failed but may be precompilable after restarting julia
15 dependencies had output during precompilation:
┌ StatsFuns
│ WARNING: Method definition (::Type{Base.MPFR.BigFloat})(Base.Irrational{:twoπ}) in module IrrationalConstants at irrationals.jl:223 overwritten in module StatsFuns on the same line (check for duplicate calls to `include`).
│ ERROR: Method overwriting is not permitted during Module precompilation. Use `__precompile__(false)` to opt-out of precompilation.
└
┌ GLMNet
│ WARNING: Method definition (::Type{Base.MPFR.BigFloat})(Base.Irrational{:twoπ}) in module IrrationalConstants at irrationals.jl:223 overwritten in module StatsFuns on the same line (check for duplicate calls to `include`).
│ ERROR: Method overwriting is not permitted during Module precompilation. Use `__precompile__(false)` to opt-out of precompilation.
└
┌ MLJ
│ WARNING: Method definition (::Type{Base.MPFR.BigFloat})(Base.Irrational{:twoπ}) in module IrrationalConstants at irrationals.jl:223 overwritten in module StatsFuns on the same line (check for duplicate calls to `include`).
│ ERROR: Method overwriting is not permitted during Module precompilation. Use `__precompile__(false)` to opt-out of precompilation.
└
┌ MLJIteration
│ WARNING: Method definition (::Type{Base.MPFR.BigFloat})(Base.Irrational{:twoπ}) in module IrrationalConstants at irrationals.jl:223 overwritten in module StatsFuns on the same line (check for duplicate calls to `include`).
│ ERROR: Method overwriting is not permitted during Module precompilation. Use `__precompile__(false)` to opt-out of precompilation.
└
┌ MLJGLMInterface
│ WARNING: Method definition (::Type{Base.MPFR.BigFloat})(Base.Irrational{:twoπ}) in module IrrationalConstants at irrationals.jl:223 overwritten in module StatsFuns on the same line (check for duplicate calls to `include`).
│ ERROR: Method overwriting is not permitted during Module precompilation. Use `__precompile__(false)` to opt-out of precompilation.
└
┌ CovarianceMatrices
│ WARNING: Method definition (::Type{Base.MPFR.BigFloat})(Base.Irrational{:twoπ}) in module IrrationalConstants at irrationals.jl:223 overwritten in module StatsFuns on the same line (check for duplicate calls to `include`).
│ ERROR: Method overwriting is not permitted during Module precompilation. Use `__precompile__(false)` to opt-out of precompilation.
└
┌ Lasso
│ WARNING: Method definition (::Type{Base.MPFR.BigFloat})(Base.Irrational{:twoπ}) in module IrrationalConstants at irrationals.jl:223 overwritten in module StatsFuns on the same line (check for duplicate calls to `include`).
│ ERROR: Method overwriting is not permitted during Module precompilation. Use `__precompile__(false)` to opt-out of precompilation.
└
┌ StatsModels
│ WARNING: Method definition (::Type{Base.MPFR.BigFloat})(Base.Irrational{:twoπ}) in module IrrationalConstants at irrationals.jl:223 overwritten in module StatsFuns on the same line (check for duplicate calls to `include`).
│ ERROR: Method overwriting is not permitted during Module precompilation. Use `__precompile__(false)` to opt-out of precompilation.
└
┌ MLJTuning
│ WARNING: Method definition (::Type{Base.MPFR.BigFloat})(Base.Irrational{:twoπ}) in module IrrationalConstants at irrationals.jl:223 overwritten in module StatsFuns on the same line (check for duplicate calls to `include`).
│ ERROR: Method overwriting is not permitted during Module precompilation. Use `__precompile__(false)` to opt-out of precompilation.
└
┌ MLJBase
│ WARNING: Method definition (::Type{Base.MPFR.BigFloat})(Base.Irrational{:twoπ}) in module IrrationalConstants at irrationals.jl:223 overwritten in module StatsFuns on the same line (check for duplicate calls to `include`).
│ ERROR: Method overwriting is not permitted during Module precompilation. Use `__precompile__(false)` to opt-out of precompilation.
└
┌ MLJModels
│ WARNING: Method definition (::Type{Base.MPFR.BigFloat})(Base.Irrational{:twoπ}) in module IrrationalConstants at irrationals.jl:223 overwritten in module StatsFuns on the same line (check for duplicate calls to `include`).
│ ERROR: Method overwriting is not permitted during Module precompilation. Use `__precompile__(false)` to opt-out of precompilation.
└
┌ MLJEnsembles
│ WARNING: Method definition (::Type{Base.MPFR.BigFloat})(Base.Irrational{:twoπ}) in module IrrationalConstants at irrationals.jl:223 overwritten in module StatsFuns on the same line (check for duplicate calls to `include`).
│ ERROR: Method overwriting is not permitted during Module precompilation. Use `__precompile__(false)` to opt-out of precompilation.
└
┌ GLM
│ WARNING: Method definition (::Type{Base.MPFR.BigFloat})(Base.Irrational{:twoπ}) in module IrrationalConstants at irrationals.jl:223 overwritten in module StatsFuns on the same line (check for duplicate calls to `include`).
│ ERROR: Method overwriting is not permitted during Module precompilation. Use `__precompile__(false)` to opt-out of precompilation.
└
┌ MLJSerialization
│ WARNING: Method definition (::Type{Base.MPFR.BigFloat})(Base.Irrational{:twoπ}) in module IrrationalConstants at irrationals.jl:223 overwritten in module StatsFuns on the same line (check for duplicate calls to `include`).
│ ERROR: Method overwriting is not permitted during Module precompilation. Use `__precompile__(false)` to opt-out of precompilation.
└
┌ Distributions
│ WARNING: Method definition (::Type{Base.MPFR.BigFloat})(Base.Irrational{:twoπ}) in module IrrationalConstants at irrationals.jl:223 overwritten in module StatsFuns on the same line (check for duplicate calls to `include`).
│ ERROR: Method overwriting is not permitted during Module precompilation. Use `__precompile__(false)` to opt-out of precompilation.
└
9 dependencies errored.
For a report of the errors see `julia> err`. To retry use `pkg> precompile`
DML function for Regression tree#
using DecisionTree
using Statistics
function DML2_for_PLM_tree(data_train, dreg, yreg; nfold=10)
nobs = nrow(data_train) # number of observations
foldid = repeat(1:nfold, ceil(Int, nobs/nfold))[shuffle(1:nobs)] # define fold indices
I = [findall(foldid .== i) for i in 1:nfold] # split observation indices into folds
ytil = fill(NaN, nobs)
dtil = fill(NaN, nobs)
println("fold: ")
for b in 1:nfold
# Exclude the current fold for training
datitanow = data_train[setdiff(1:nobs, I[b]), Not(:d)]
datitanoy = data_train[setdiff(1:nobs, I[b]), Not(:y)]
# Current fold for prediction
datitanowpredict = data_train[I[b], Not(:d)]
datitanoypredict = data_train[I[b], Not(:y)]
# Fit models
dfit = dreg(datitanoy)
yfit = yreg(datitanow)
# Make predictions
dhat = predict(dfit, DataFrame(datitanoypredict, :auto))
yhat = predict(yfit, DataFrame(datitanowpredict, :auto))
# Record residuals
dtil[I[b]] .= data_train[I[b], :d] .- dhat
ytil[I[b]] .= data_train[I[b], :y] .- yhat
print("$b ")
end
# Ensure ytil and dtil are column vectors
ytil = reshape(ytil, :, 1)
dtil = reshape(dtil, :, 1)
# Regress one residual on the other
rfit = lm(@formula(ytil ~ dtil), DataFrame(hcat(ytil, dtil), :auto))
coef_est = coef(rfit)[2]
se = sqrt(vcov(HC0, rfit)[2, 2])
println("\ncoef (se) = $coef_est ($se)")
return (coef_est = coef_est, se = se, dtil = dtil, ytil = ytil)
end
DML2_for_PLM_tree (generic function with 1 method)
DML function for Boosting Trees#
using MLJ
using XGBoost
function DML2_for_PLM_boosttree(data_train, dreg, yreg; nfold=10)
nobs = nrow(data_train) # number of observations
foldid = repeat(1:nfold, ceil(Int, nobs/nfold))[shuffle(1:nobs)] # define fold indices
I = [findall(foldid .== i) for i in 1:nfold] # split observation indices into folds
ytil = fill(NaN, nobs)
dtil = fill(NaN, nobs)
println("fold: ")
for b in 1:nfold
# Exclude the current fold for training
datitanow = data_train[setdiff(1:nobs, I[b]), Not(:d)]
datitanoy = data_train[setdiff(1:nobs, I[b]), Not(:y)]
# Current fold for prediction
datitanowpredict = data_train[I[b], Not(:d)]
datitanoypredict = data_train[I[b], Not(:y)]
# Fit models
dfit = dreg(datitanoy)
best_boostt = fit!(dfit, verbosity=0) # fit the model
yfit = yreg(datitanow)
best_boosty = fit!(yfit, verbosity=0) # fit the model
# Make predictions
dhat = MLJ.predict(best_boostt, DataFrame(datitanoypredict, :auto))
yhat = MLJ.predict(best_boosty, DataFrame(datitanowpredict, :auto))
# Record residuals
dtil[I[b]] .= data_train[I[b], :d] .- dhat
ytil[I[b]] .= data_train[I[b], :y] .- yhat
print("$b ")
end
# Ensure ytil and dtil are column vectors
ytil = reshape(ytil, :, 1)
dtil = reshape(dtil, :, 1)
# Regress one residual on the other
rfit = lm(@formula(ytil ~ dtil), DataFrame(hcat(ytil, dtil), :auto))
coef_est = coef(rfit)[2]
se = sqrt(vcov(HC0, rfit)[2, 2])
println("\ncoef (se) = $coef_est ($se)")
return (coef_est = coef_est, se = se, dtil = dtil, ytil = ytil)
end
DML2_for_PLM_boosttree (generic function with 1 method)
DML function for Lasso#
using DataFrames
using CSV
using GLM
using Random
using StatsBase
using GLMNet
using Statistics
function DML2_for_PLM(x, d, y, dreg, yreg; nfold=10)
nobs = size(x, 1) # number of observations
foldid = repeat(1:nfold, ceil(Int, nobs/nfold))[shuffle(1:nobs)] # define fold indices
I = [findall(foldid .== i) for i in 1:nfold] # split observation indices into folds
ytil = fill(NaN, nobs)
dtil = fill(NaN, nobs)
println("fold: ")
for b in 1:nfold
# Exclude the current fold for training
train_indices = setdiff(1:nobs, I[b])
test_indices = I[b]
# Fit models
dfit = dreg(x[train_indices, :], d[train_indices])
yfit = yreg(x[train_indices, :], y[train_indices])
# Make predictions
dhat = GLMNet.predict(dfit, x[test_indices, :])
yhat = GLMNet.predict(yfit, x[test_indices, :])
# Record residuals
dtil[test_indices] .= d[test_indices] .- dhat
ytil[test_indices] .= y[test_indices] .- yhat
print("$b ")
end
# Ensure ytil and dtil are column vectors
ytil = vec(ytil)
dtil = vec(dtil)
# Regress one residual on the other
data = DataFrame(ytil = ytil, dtil = dtil)
rfit = lm(@formula(ytil ~ dtil), data)
coef_est = coef(rfit)[2]
se = sqrt(vcov(HC0, rfit)[2, 2])
println("\ncoef (se) = $coef_est ($se)")
return Dict("coef.est" => coef_est, "se" => se, "dtil" => dtil, "ytil" => ytil)
end
DML2_for_PLM (generic function with 1 method)
3.1. Lasso#
# Define Lasso regression functions
dreg_lasso = (x, d) -> GLMNet.glmnet(x, d, alpha=1.0, lambda=[0.1])
yreg_lasso = (x, y) -> GLMNet.glmnet(x, y, alpha=1.0, lambda=[0.1])
# Apply DML with Lasso
DML2_lasso = DML2_for_PLM(x, d, y, dreg_lasso, yreg_lasso, nfold=10)
# Extract results
coef_lasso = DML2_lasso["coef.est"]
se_lasso = DML2_lasso["se"]
prRes_lassoD = mean((DML2_lasso["dtil"]).^2)
prRes_lassoY = mean((DML2_lasso["ytil"]).^2)
# Format results
prRes_lasso = DataFrame(
Estimate = coef_lasso,
`Standard Error` = se_lasso,
`RMSE D` = sqrt(prRes_lassoD),
`RMSE Y` = sqrt(prRes_lassoY)
)
# Display the results
println(prRes_lasso)
fold:
1 2 3 4 5 6 7 8 9 10
UndefVarError: `coef` not defined
Stacktrace:
[1] DML2_for_PLM(x::Matrix{Float64}, d::Vector{Float64}, y::Vector{Float64}, dreg::var"#18#19", yreg::var"#20#21"; nfold::Int64)
@ Main .\In[8]:43
[2] top-level scope
@ In[9]:8
The message treatment providing information about Internet-accessed sexually transmitted infection testing predicts an increase in the probability that a person will get tested by 25.13 percentage points compared to receiving information about nearby clinics offering in-person testing. By providing both groups with information about testing, we mitigate the potential reminder effect, as both groups are equally prompted to consider testing. This approach allows us to isolate the impact of the type of information “Internet-accessed testing” versus “in-person clinic testing” on the likelihood of getting tested. Through randomized assignment, we establish causality rather than mere correlation, confirming that the intervention’s effect is driven by the unique advantages of Internet-accessed testing, such as increased privacy, reduced embarrassment, and convenience
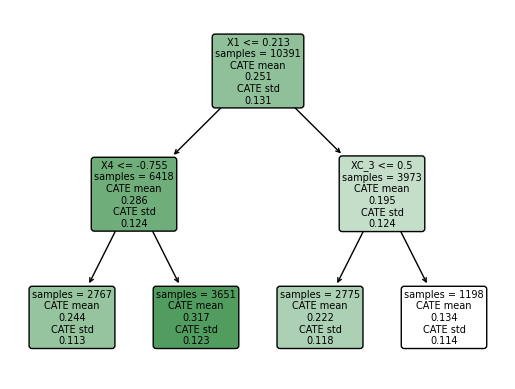
3.2. Regression Trees#
# Set up the basic formula for the regression tree
X_basic = "gender_transgender + ethnicgrp_asian + ethnicgrp_black + ethnicgrp_mixed_multiple+ ethnicgrp_other + ethnicgrp_white + partners1 + postlaunch + msm + age+ imd_decile"
y_form_tree = @formula(y ~ $(Meta.parse(X_basic)))
t_form_tree = @formula(w ~ $(Meta.parse(X_basic)))
# Define tree regression functions
yreg_tree = (dataa) -> DecisionTreeRegressor(min_samples_leaf=5, max_depth=5).fit(Matrix(dataa[:, Not(:y)]), Vector(dataa[:, :y]))
treg_tree = (dataa) -> DecisionTreeRegressor(min_samples_leaf=5, max_depth=5).fit(Matrix(dataa[:, Not(:w)]), Vector(dataa[:, :w]))
# Apply DML with regression tree
DML2_tree = DML2_for_PLM_tree(data_train, treg_tree, yreg_tree, nfold=10)
# Extract results
coef_tree = DML2_tree[1]
se_tree = DML2_tree[2]
prRes_treeD = mean((DML2_tree[3]).^2)
prRes_treeY = mean((DML2_tree[4]).^2)
# Format results
prRes_tree = DataFrame(
Estimate = coef_tree,
`Standard Error` = se_tree,
`RMSE D` = sqrt(prRes_treeD),
`RMSE Y` = sqrt(prRes_treeY)
)
# Display the results
println(prRes_tree)
LoadError: ArgumentError: interpolation with $ not supported in @formula. Use @eval @formula(...) instead.
in expression starting at In[23]:4
Stacktrace:
[1] catch_dollar
@ C:\Users\juanl\.julia\packages\StatsModels\fK0P3\src\formula.jl:20 [inlined]
[2] parse!(ex::Expr, rewrites::Vector{DataType})
@ StatsModels C:\Users\juanl\.julia\packages\StatsModels\fK0P3\src\formula.jl:180
[3] parse!(ex::Expr, rewrites::Vector{DataType})
@ StatsModels C:\Users\juanl\.julia\packages\StatsModels\fK0P3\src\formula.jl:197
[4] parse!(x::Expr)
@ StatsModels C:\Users\juanl\.julia\packages\StatsModels\fK0P3\src\formula.jl:176
[5] var"@formula"(__source__::LineNumberNode, __module__::Module, ex::Any)
@ StatsModels C:\Users\juanl\.julia\packages\StatsModels\fK0P3\src\formula.jl:63
The message treatment providing information about Internet-accessed sexually transmitted infection testing predicts an increase in the probability that a person will get tested by 23.08 percentage points compared to receiving information about nearby clinics offering in-person testing. By providing both groups with information about testing, we mitigate the potential reminder effect, as both groups are equally prompted to consider testing. This approach allows us to isolate the impact of the type of information “Internet-accessed testing” versus “in-person clinic testing” on the likelihood of getting tested. Through randomized assignment, we establish causality rather than mere correlation, confirming that the intervention’s effect is driven by the unique advantages of Internet-accessed testing, such as increased privacy, reduced embarrassment, and convenience
3.3. Boosting Trees#
# Set up the basic formula for the boosted tree
X_basic = "gender_transgender + ethnicgrp_asian + ethnicgrp_black + ethnicgrp_mixed_multiple+ ethnicgrp_other + ethnicgrp_white + partners1 + postlaunch + msm + age+ imd_decile"
y_form_tree = @formula(y ~ $(Meta.parse(X_basic)))
t_form_tree = @formula(w ~ $(Meta.parse(X_basic)))
# Define boosted tree regression functions
yreg_treeboost = (dataa) -> machine(XGBoostRegressor(max_depth=2, nrounds=1000, eta=0.01, subsample=0.5), dataa[:, Not(:y)], dataa[:, :y])
treg_treeboost = (dataa) -> machine(XGBoostRegressor(max_depth=2, nrounds=1000, eta=0.01, subsample=0.5), dataa[:, Not(:w)], dataa[:, :w])
# Apply DML with boosted trees
DML2_boosttree = DML2_for_PLM_boosttree(data_train, treg_treeboost, yreg_treeboost, nfold=10)
# Extract results
coef_boosttree = DML2_boosttree[1]
se_boosttree = DML2_boosttree[2]
prRes_boosttreeD = mean((DML2_boosttree[3]).^2)
prRes_boosttreeY = mean((DML2_boosttree[4]).^2)
# Format results
prRes_boosttree = DataFrame(
Estimate = coef_boosttree,
`Standard Error` = se_boosttree,
`RMSE D` = sqrt(prRes_boosttreeD),
`RMSE Y` = sqrt(prRes_boosttreeY)
)
# Display the results
println(prRes_boosttree)
LoadError: ArgumentError: interpolation with $ not supported in @formula. Use @eval @formula(...) instead.
in expression starting at In[25]:3
Stacktrace:
[1] catch_dollar
@ C:\Users\juanl\.julia\packages\StatsModels\fK0P3\src\formula.jl:20 [inlined]
[2] parse!(ex::Expr, rewrites::Vector{DataType})
@ StatsModels C:\Users\juanl\.julia\packages\StatsModels\fK0P3\src\formula.jl:180
[3] parse!(ex::Expr, rewrites::Vector{DataType})
@ StatsModels C:\Users\juanl\.julia\packages\StatsModels\fK0P3\src\formula.jl:197
[4] parse!(x::Expr)
@ StatsModels C:\Users\juanl\.julia\packages\StatsModels\fK0P3\src\formula.jl:176
[5] var"@formula"(__source__::LineNumberNode, __module__::Module, ex::Any)
@ StatsModels C:\Users\juanl\.julia\packages\StatsModels\fK0P3\src\formula.jl:63
The message treatment providing information about Internet-accessed sexually transmitted infection testing predicts an increase in the probability that a person will get tested by 25.28 percentage points compared to receiving information about nearby clinics offering in-person testing. By providing both groups with information about testing, we mitigate the potential reminder effect, as both groups are equally prompted to consider testing. This approach allows us to isolate the impact of the type of information “Internet-accessed testing” versus “in-person clinic testing” on the likelihood of getting tested. Through randomized assignment, we establish causality rather than mere correlation, confirming that the intervention’s effect is driven by the unique advantages of Internet-accessed testing, such as increased privacy, reduced embarrassment, and convenience
3.4. Random Forest#
# Function for Double Machine Learning with Partially Linear Model using Random Forest
function DML2_for_PLM_RF(data_train, dreg, yreg; nfold=10)
nobs = nrow(data_train) # number of observations
foldid = repeat(1:nfold, ceil(Int, nobs/nfold))[shuffle(1:nobs)] # define fold indices
I = [findall(foldid .== i) for i in 1:nfold] # split observation indices into folds
ytil = fill(NaN, nobs)
dtil = fill(NaN, nobs)
println("fold: ")
for b in 1:nfold
# Exclude the current fold for training
train_indices = setdiff(1:nobs, I[b])
test_indices = I[b]
# Fit models
dfit = dreg(x[train_indices, :], d[train_indices])
yfit = yreg(x[train_indices, :], y[train_indices])
# Make predictions
dhat = predict(dfit, x[test_indices, :])
yhat = predict(yfit, x[test_indices, :])
# Record residuals
dtil[test_indices] .= d[test_indices] .- dhat
ytil[test_indices] .= y[test_indices] .- yhat
print("$b ")
end
# Ensure ytil and dtil are column vectors
ytil = vec(ytil)
dtil = vec(dtil)
# Regress one residual on the other
data = DataFrame(ytil = ytil, dtil = dtil)
rfit = lm(@formula(ytil ~ dtil), data)
coef_est = coef(rfit)[2]
se = sqrt(vcov(HC0, rfit)[2, 2])
println("\ncoef (se) = $coef_est ($se)")
return Dict("coef.est" => coef_est, "se" => se, "dtil" => dtil, "ytil" => ytil)
end
DML2_for_PLM_RF (generic function with 1 method)
# Define Random Forest regression functions
dreg_RF = (x, d) -> RandomForestRegressor(n_trees=100).fit(x, d)
yreg_RF = (x, y) -> RandomForestRegressor(n_trees=100).fit(x, y)
# Apply DML with Random Forest
DML2_RF = DML2_for_PLM_RF(x, d, y, dreg_RF, yreg_RF, nfold=10)
# Extract results
coef_RF = DML2_RF["coef.est"]
se_RF = DML2_RF["se"]
prRes_RFD = mean((DML2_RF["dtil"]).^2)
prRes_RFY = mean((DML2_RF["ytil"]).^2)
# Format results
prRes_RF = DataFrame(
Estimate = coef_RF,
`Standard Error` = se_RF,
`RMSE D` = sqrt(prRes_RFD),
`RMSE Y` = sqrt(prRes_RFY)
)
# Display the results
println(prRes_RF)
MethodError: no method matching DML2_for_PLM_RF(::Matrix{Float64}, ::Vector{Float64}, ::Vector{Float64}, ::var"#55#56", ::var"#57#58"; nfold::Int64)
Closest candidates are:
DML2_for_PLM_RF(::Any, ::Any, ::Any; nfold)
@ Main In[26]:2
Stacktrace:
[1] top-level scope
@ In[27]:7
The message treatment providing information about Internet-accessed sexually transmitted infection testing predicts an increase in the probability that a person will get tested by 24.14 percentage points compared to receiving information about nearby clinics offering in-person testing. By providing both groups with information about testing, we mitigate the potential reminder effect, as both groups are equally prompted to consider testing. This approach allows us to isolate the impact of the type of information “Internet-accessed testing” versus “in-person clinic testing” on the likelihood of getting tested. Through randomized assignment, we establish causality rather than mere correlation, confirming that the intervention’s effect is driven by the unique advantages of Internet-accessed testing, such as increased privacy, reduced embarrassment, and convenience
3.5. Table and Coefficient plot#
Table#
Coefficient Plot#
prRes_D = [
mean((DML2_lasso["dtil"]).^2),
mean((DML2_tree["dtil"]).^2),
mean((DML2_boosttree["dtil"]).^2),
mean((DML2_RF["dtil"]).^2)
]
prRes_Y = [
mean((DML2_lasso["ytil"]).^2),
mean((DML2_tree["ytil"]).^2),
mean((DML2_boosttree["ytil"]).^2),
mean((DML2_RF["ytil"]).^2)
]
prRes = hcat(sqrt.(prRes_D), sqrt.(prRes_Y))
rownames!(prRes, ["RMSE D", "RMSE Y"])
colnames!(prRes, ["Lasso", "Reg Tree", "Boost Tree", "Random Forest"])
# Create results table
table = zeros(4, 4)
# Point Estimate
table[1, 1] = DML2_lasso["coef.est"]
table[2, 1] = DML2_tree["coef.est"]
table[3, 1] = DML2_boosttree["coef.est"]
table[4, 1] = DML2_RF["coef.est"]
# SE
table[1, 2] = DML2_lasso["se"]
table[2, 2] = DML2_tree["se"]
table[3, 2] = DML2_boosttree["se"]
table[4, 2] = DML2_RF["se"]
# RMSE Y
table[1, 3] = prRes[2, 1]
table[2, 3] = prRes[2, 2]
table[3, 3] = prRes[2, 3]
table[4, 3] = prRes[2, 4]
# RMSE D
table[1, 4] = prRes[1, 1]
table[2, 4] = prRes[1, 2]
table[3, 4] = prRes[1, 3]
table[4, 4] = prRes[1, 4]
# Print results
colnames!(table, ["Estimate", "Standard Error", "RMSE Y", "RMSE D"])
rownames!(table, ["Lasso", "Reg Tree", "Boost Tree", "Random Forest"])
# Display the table
println(table)
UndefVarError: `DML2_lasso` not defined
Stacktrace:
[1] top-level scope
@ In[28]:2
using Pkg
Pkg.add("Plots")
using plots
Resolving package versions...
No Changes to `C:\Users\juanl\.julia\environments\v1.10\Project.toml`
No Changes to `C:\Users\juanl\.julia\environments\v1.10\Manifest.toml`
Precompiling project...
? StatsFuns
? StatsModels
? Distributions
? GLM
? CovarianceMatrices
? GLMNet
? MLJGLMInterface
? MLJBase
? Lasso
? MLJIteration
? MLJTuning
? MLJEnsembles
? MLJSerialization
? MLJModels
? MLJ
ArgumentError: Package plots not found in current path.
- Run `import Pkg; Pkg.add("plots")` to install the plots package.
Stacktrace:
[1] macro expansion
@ .\loading.jl:1772 [inlined]
[2] macro expansion
@ .\lock.jl:267 [inlined]
[3] __require(into::Module, mod::Symbol)
@ Base .\loading.jl:1753
[4] #invoke_in_world#3
@ .\essentials.jl:926 [inlined]
[5] invoke_in_world
@ .\essentials.jl:923 [inlined]
[6] require(into::Module, mod::Symbol)
@ Base .\loading.jl:1746
# Convert `table` to DataFrame
table_ci = DataFrame(table, ["Estimate", "Standard Error", "RMSE Y", "RMSE D"])
table_ci.Method = ["Lasso", "Reg Tree", "Boost Tree", "Random Forest"]
# Calculate confidence intervals
table_ci.CI_Lower_1 = table_ci.Estimate .- 2.576 .* table_ci."Standard Error"
table_ci.CI_Upper_1 = table_ci.Estimate .+ 2.576 .* table_ci."Standard Error"
table_ci.CI_Lower_5 = table_ci.Estimate .- 1.96 .* table_ci."Standard Error"
table_ci.CI_Upper_5 = table_ci.Estimate .+ 1.96 .* table_ci."Standard Error"
table_ci.CI_Lower_10 = table_ci.Estimate .- 1.645 .* table_ci."Standard Error"
table_ci.CI_Upper_10 = table_ci.Estimate .+ 1.645 .* table_ci."Standard Error"
# Plotting
plot(table_ci.Method, table_ci.Estimate, seriestype=:scatter, label="Estimate", markersize=4)
plot!(table_ci.Method, table_ci.CI_Lower_5, ribbon=(table_ci.CI_Upper_5 .- table_ci.CI_Lower_5), label="95% CI", lw=2, lc=:blue, alpha=0.7)
plot!(table_ci.Method, table_ci.CI_Lower_10, ribbon=(table_ci.CI_Upper_10 .- table_ci.CI_Lower_10), label="90% CI", lw=2, lc=:red, alpha=0.7)
plot!(table_ci.Method, table_ci.CI_Lower_1, ribbon=(table_ci.CI_Upper_1 .- table_ci.CI_Lower_1), label="99% CI", lw=2, lc=:green, alpha=0.7)
title!("Estimated Coefficients with Confidence Intervals")
xlabel!("Method")
ylabel!("Estimate")
theme(:minimal)
xticks!(1:4, table_ci.Method)
xrotation!(45)
MethodError: no method matching DataFrame(::typeof(table), ::Vector{String})
Closest candidates are:
DataFrame(::AbstractVector{<:Type}, ::AbstractVector{<:AbstractString}, ::Integer; makeunique)
@ DataFrames C:\Users\juanl\.julia\packages\DataFrames\JZ7x5\src\dataframe\dataframe.jl:397
DataFrame(::AbstractVector{<:AbstractVector}, ::AbstractVector; makeunique, copycols)
@ DataFrames C:\Users\juanl\.julia\packages\DataFrames\JZ7x5\src\dataframe\dataframe.jl:344
DataFrame(::AbstractVector{<:Type}, ::AbstractVector{<:AbstractString}; ...)
@ DataFrames C:\Users\juanl\.julia\packages\DataFrames\JZ7x5\src\dataframe\dataframe.jl:397
...
Stacktrace:
[1] top-level scope
@ In[37]:2
3.6. Model#
To choose the best model, we must compare the RMSEs of the outcome variable Y. In this case, the model with the lowest RMSE for Y is generated by Lasso (0.4716420), whereas the lowest for the treatment is generated by Boosting Trees (0.4983734). Therefore, DML could be employed with Y cleaned using Lasso and the treatment using Boosting Trees.