Lab 3 - R Code#
Group 3: Valerie Dube, Erzo Garay, Juan Marcos Guerrero y Matías Villalba,
1. Neyman Orthogonality Proof#
\(Y_{nx1}=\alpha D_{nx1}+W_{nxp}\beta_{px1}+\epsilon_{nx1}\\ \widetilde{Y}_{nx1}=Y_{nx1}-W_{nxp}X_{{{Yw}_{px1}}}\\ \widetilde{D}_{nx1}=D_{nx1}-W_{nxp}X_{{{Dw}_{px1}}}\\ W=Matrix\ composed\ by\ one\ variable\ in\ each\ column\\ X=Vector\ of\ coefficients\\ n=N° observations\\ p=N° confounders \\\ \\\ \\\ M(a,\eta)=E[(\widetilde{Y}_{nx1}(\eta_{1})-a\widetilde{D}(\eta_{2})_{nx1})'(\widetilde{D}(\eta_{2})_{nx1})]=0\\ \frac{\partial a}{\partial \eta}=-[\frac{\partial M}{\partial a}(\alpha,\eta_{0})]^{-1}[\frac{\partial M}{\partial \eta}(\alpha,\eta_{0})]\\ \frac{\partial M}{\partial \eta}(\alpha,\eta_{0})=\frac{\partial M}{\partial \eta_{1}}(\alpha,X_{Yw},X_{Dw})+\frac{\partial M}{\partial \eta_{2}}(\alpha,X_{Yw},X_{Dw})\\ S_{1}=\frac{\partial M}{\partial \eta_{1}}(\alpha,X_{Yw},X_{Dw}) , S_{2}=\frac{\partial M}{\partial \eta_{2}}(\alpha,X_{Yw},X_{Dw})\)
Demonstration 1:#
\(Given\ the\ general\ formula\,\ before\ using\ (\alpha,X_{Yw},X_{Dw}): \widetilde{Y}_{nx1}=Y_{nx1}-W_{nxp}\eta_{{{1}_{px1}}} ,\ \widetilde{D}_{nx1}=D_{nx1}-W_{nxp}\eta_{{{2}_{px1}}}: \\\ \\\ S_{1}=\frac{\partial M}{\partial \eta_{1}}|_{(\alpha,W_{Yw},W_{Dw})}\\\ \\\ =\frac{\partial E[(\widetilde{Y}_{nx1}(\eta_{1})-a\widetilde{D}(\eta_{2})_{nx1})'(\widetilde{D}(\eta_{2})_{nx1})}{\partial \eta_{1}}|_{(\alpha,W_{Yw},W_{Dw})}=\frac{\partial E[Y_{nx1}-W_{nxp}\eta_{{{1}_{px1}}}-aD_{nx1}+aW_{nxp}\eta_{{{2}_{px1}}})'(D_{nx1}-W_{nxp}\eta_{{{2}_{px1}}})}{\partial \eta_{1}}|_{(\alpha,W_{Yw},W_{Dw})}\\\ \\\ Substituting\ (\alpha,W_{Yw},W_{Dw})\ in\ (a,\eta_{1},\eta_{2})\\\ \\\ =\frac{\partial E[(\widetilde{Y}_{nx1}(\eta_{1})-a\widetilde{D}(\eta_{2})_{nx1})'(\widetilde{D}(\eta_{2})_{nx1})]}{\partial \eta_{1}}|_{(\alpha,W_{Yw},W_{Dw})}=E[(W_{nxp})'(D_{nx1}-W_{n*p}\eta_{{2}_{px1}})]|_{(\alpha,W_{Yw},W_{Dw})}\\ =E[(W_{nxp})'(D_{nx1}-W_{n*p}X_{{{Dw}_{px1}}})]\\ =E[W'_{pxn}D_{nx1}-W'_{pxn}W_{nxp}(W'_{pxn}W_{nxp})^{-1}(W'_{pxn}D_{nx1})]\\ =E[W'_{pxn}D_{nx1}-I_{pxp}(W'_{pxn}D_{nx1})]=E[W'_{pxn}D_{nx1}-W'_{pxn}D_{nx1}]=E[0]\\ S_{1}=0\)
Demonstration 2:#
\(Given\ the\ general\ formula\,\ before\ using\ (\alpha,X_{Yw},X_{Dw}): \widetilde{Y}_{nx1}=y_{nx1}-W_{nxp}\eta_{{{1}_{px1}}} ,\ \widetilde{D}_{nx1}=D_{nx1}-W_{nxp}\eta_{{{2}_{px1}}}\\\ \\\ S_{2}=\frac{\partial M}{\partial \eta_{2}}|_{(\alpha,W_{Yw},W_{Dw})}=0\\\ \\ =\frac{\partial E[(\widetilde{Y}_{nx1}(\eta_{1})-a\widetilde{D}(\eta_{2})_{nx1})'(\widetilde{D}(\eta_{2})_{nx1})}{\partial \eta_{2}}|_{(\alpha,W_{Yw},W_{Dw})}=\frac{\partial E[Y_{nx1}-W_{nxp}\eta_{{{1}_{px1}}}-aD_{nx1}+aW_{nxp}\eta_{{{2}_{px1}}})'(D_{nx1}-W_{nxp}\eta_{{{2}_{px1}}})}{\partial \eta_{2}}|_{(\alpha,W_{Yw},W_{Dw})}\\ =\frac{\partial E[-Y_{1xn}'W_{nxp}\eta_{{2}_{px1}}+\eta'_{{1}_{1xn}}W'_{pxn}W_{pxn}\eta_{{2}_{px1}}+aD'_{1xn}W_{nxp}\eta_{{2}_{px1}}+a\eta'_{{2}_{1xp}}W'_{pxn}D_{nx1}-a\eta'_{{2}_{1xp}}W'_{pxn}W_{nxp}\eta_{{2}_{px1}}]}{\partial \eta_{2}}|_{(\alpha,W_{Yw},W_{Dw})}\\ =E[-W'_{pxn}Y_{nx1}+W'_{pxn}W_{nxp}\eta_{{1}_{px1}}+aW'_{pxn}D_{nx1}+aW'_{pxn}D_{nx1}-aW'_{pxn}W_{nxp}\eta_{{2}_{px1}}-aW'_{pxn}W_{nxp}\eta_{{2}_{px1}}]|_{(\alpha,W_{Yw},W_{Dw})}\\\ \\\ Substituting\ (\alpha,W_{Yw},W_{Dw})\ in\ (a,\eta_{1},\eta_{2})\\\ \\ =E[-W'_{pxn}Y_{nx1}+W'_{pxn}W_{nxp}X_{{{yw}_{px1}}}+\alpha W'_{pxn}D_{nx1}+\alpha W'_{pxn}D_{nx1}-\alpha W'_{pxn}W_{nxp}X_{{{Dw}_{px1}}}-\alpha W'_{pxn}W_{nxp}X_{{{Dw}_{px1}}}]\\ =E[-W'_{pxn}Y_{nx1}+W'_{pxn}W_{nxp}(W'_{pxn}W_{nxp})^{-1}(W'_{pxn}Y_{nx1})+\alpha W'_{pxn}D_{nx1}+\alpha W'_{pxn}D_{nx1}-\alpha W'_{pxn}W_{nxp}(W'_{pxn}W_{nxp})^{-1}(W'_{pxn}D_{nx1})-\alpha W'_{pxn}W_{nxp}(W'_{pxn}W_{nxp})^{-1}(W'_{pxn}D_{nx1})]\\ =E[-W'_{pxn}Y_{nx1}+-W'_{pxn}Y_{nx1}+\alpha W'_{pxn}D_{nx1}+\alpha W'_{pxn}D_{nx1}-\alpha W'_{pxn}D_{nx1}-\alpha W'_{pxn}D_{nx1}]=E[0]=0\\ S_{2}=0\)
2. Code Section#
2.1. Orthogonal Learning#
# Install and load necessary packages
#install.packages(c("glmnet", "parallel", "ggplot2", "gridExtra", "hdm", "cowplot"))
library(hdm)
library(glmnet)
library(parallel)
library(ggplot2)
library(gridExtra)
library(cowplot)
Loading required package: Matrix
Loaded glmnet 4.1-8
Simulation Design#
We are going to simulate 3 different trials to show the properties we talked about orthogonal learning.
For that we first define a function that runs a single observation of our simulation.
simulate_once <- function(seed) {
set.seed(seed)
n <- 100
p <- 100
beta <- 1 / (1:p)^2
gamma <- 1 / (1:p)^2
mean <- 0
sd <- 1
X <- matrix(rnorm(n * p, mean, sd), n, p)
D <- (X %*% gamma) + rnorm(n, mean, sd) / 4
Y <- 10 * D + (X %*% beta) + rnorm(n, mean, sd)
# Single selection method
r_lasso <- rlasso(Y ~ D + X, post = TRUE)
coef_matrix <- r_lasso$coefficients[-c(1, 2)]
SX_IDs <- which(coef_matrix != 0)
# In case all X coefficients are zero
if (length(SX_IDs) == 0) {
naive_model <- lm(Y ~ D)
naive_coef <- coef(summary(naive_model))[2, 1]
} else {
X_D <- cbind(D, X[, SX_IDs])
naive_model <- lm(Y ~ X_D)
naive_coef <- coef(summary(naive_model))[2, 1]
}
# Regress residuals
resY <- residuals(rlasso(Y ~ X, post = FALSE))
resD <- residuals(rlasso(D ~ X, post = FALSE))
orthogonal_model <- lm(resY ~ resD)
orthogonal_coef <- coef(summary(orthogonal_model))[2, 1]
return(c(naive_coef, orthogonal_coef))
}
Then we define a function that runs the simulation on its enterity, using parallel computing and the function we previously defined.
run_simulation <- function(B) {
cl <- makeCluster(detectCores()) # Create a cluster using all available cores
on.exit({
stopCluster(cl)
}, add = TRUE) # Ensure the cluster is stopped when the function exits, even if there's an error
clusterExport(cl, list("simulate_once", "rlasso", "coef", "residuals", "summary", "set.seed", "matrix", "rnorm", "cbind", "lm", "which")) # Export necessary objects and functions to the cluster
results <- tryCatch({
parLapply(cl, 1:B, simulate_once) # Run the simulation in parallel
}, error = function(e) {
message("Error during parallel execution: ", e)
stopCluster(cl)
stop(e)
})
Naive <- sapply(results, function(x) x[1])
Orthogonal <- sapply(results, function(x) x[2])
return(list(Naive = Naive, Orthogonal = Orthogonal))
}
Orto_breaks <- seq(8, 12, by = 0.2)
Naive_breaks <- seq(8, 12, by = 0.2)
Next we run the simulations with 100, 1000, and 10000 iterations and plot the histograms for the Naive and Orthogonal estimations
options(warn=-1)
Bs <- c(100, 1000, 10000)
custom_theme <- theme_classic() +
theme(
panel.background = element_blank(),
panel.border = element_rect(color = "black", fill = NA, linewidth = 1),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
plot.background = element_blank()
)
for (B in Bs) {
start_time <- Sys.time()
results <- run_simulation(B)
Naive <- results$Naive
Orthogonal <- results$Orthogonal
end_time <- Sys.time()
elapsed_time <- as.numeric(difftime(end_time, start_time, units = "secs"))
df1 <- data.frame(Orthogonal = Orthogonal)
df2 <- data.frame(Naive = Naive)
vlines_df <- data.frame(x = c(mean(df1$Orthogonal), 10),
label = c("Estimated Treatment Effect", "True Effect"))
p1 <- ggplot(df1, aes(x = Orthogonal)) +
geom_histogram(aes(y = after_stat(density)), breaks = Orto_breaks, fill = "lightblue", color = "NA") +
geom_density(color = "darkblue", linewidth=1) +
geom_vline(data = vlines_df, aes(xintercept = x, color = label, linetype = label), linewidth = 1) +
scale_x_continuous(limits = c(8, 12)) +
ggtitle("Orthogonal estimation") +
xlab("Estimated Treatment Effect")+
scale_color_manual(values = c("Estimated Treatment Effect" = "blue", "True Effect" = "red")) +
scale_linetype_manual(values = c("Estimated Treatment Effect" = "dashed", "True Effect" = "dashed")) +
custom_theme+
labs(color = "Legend", linetype = "Legend")+
theme(legend.direction = "horizontal")
p2 <- ggplot(df2, aes(x = Naive)) +
geom_histogram(aes(y = after_stat(density)), breaks = Naive_breaks, fill = "lightblue", color = "NA") +
geom_density(color = "darkblue", linewidth=1) +
geom_vline(aes(xintercept = mean(Naive)), color = "blue", linetype = "dashed", linewidth=1) +
geom_vline(xintercept = 10, color = "red", linetype = "dashed", linewidth=1) +
scale_x_continuous(limits = c(8, 12)) +
ggtitle("Naive estimation") +
xlab("Estimated Treatment Effect")+
custom_theme
# Extract the legend
legend <- suppressWarnings(get_legend(p1))
# Remove the legends from the individual plots
p1 <- p1 + theme(legend.position = "none")
p2 <- p2 + theme(legend.position = "none")
# Arrange the plots and the legend
combined_plot <- plot_grid(p1, p2, ncol = 2, align = "v")
combined_plot_with_legend <- plot_grid(combined_plot, legend, ncol = 1, rel_heights = c(1, 0.07))
grid.arrange(combined_plot_with_legend, ncol = 1, top = paste("Simulation with", B, "iterations"))
print(paste("Time taken for", B, "iteration simulation:", round(elapsed_time, 2), "seconds"))
}
[1] "Time taken for 100 iteration simulation: 4.61 seconds"
We can se that the orthogonal estimation yields on average a coeficient more centered on the true effect than the naive estimation. This method of estimation that utilizes the residuals of lasso regresion and helps reduce bias more efficiently than the naive estimation method. This is because it leverages the properties explained on the begining of this notebook rather than just controlling for a relevant set of covariates which may induce endogeneity.
We have used parallel computing because it is supposed to allow us to achieve better processing times. It can significantly lower the running time of computations due to its ability to distribute the workload across multiple processing units.
As an example, we can see how long it would have taken to run the simulation with 1000 iteration if we had not utilized parallel computing.
Without parallel computing
run_simulation <- function(B) {
Naive <- numeric(B)
Orthogonal <- numeric(B)
for (i in 1:B) {
results <- simulate_once(i)
Naive[i] <- results[1]
Orthogonal[i] <- results[2]
}
return(list(Naive = Naive, Orthogonal = Orthogonal))
}
start_time <- Sys.time()
results <- run_simulation(1000)
Naive <- results$Naive
Orthogonal <- results$Orthogonal
end_time <- Sys.time()
elapsed_time <- as.numeric(difftime(end_time, start_time, units = "secs"))
print(paste("Time taken for 1000 iteration simulation:", round(elapsed_time, 2), "seconds"))
[1] "Time taken for 1000 iteration simulation: 129.2 seconds"
We can see that if we were to not use parallel computing, the processing time would be higher. It took 129 seconds to achieve what we achieved in 27.
2.2. Double Lasso - Using School data#
# Install necessary packages
install.packages("tidyverse")
install.packages("lme4")
install.packages("stargazer")
install.packages("caTools")
install.packages("caret")
install.packages("glmnet")
Warning message:
"dependencies 'MASS', 'Matrix', 'lattice' are not available"also installing the dependencies 'fastmap', 'colorspace', 'sys', 'bit', 'ps', 'sass', 'cachem', 'nlme', 'farver', 'munsell', 'rappdirs', 'askpass', 'bit64', 'processx', 'highr', 'xfun', 'yaml', 'bslib', 'fontawesome', 'htmltools', 'tinytex', 'backports', 'ellipsis', 'glue', 'lifecycle', 'memoise', 'blob', 'tidyselect', 'vctrs', 'data.table', 'gtable', 'isoband', 'mgcv', 'scales', 'gargle', 'cellranger', 'curl', 'ids', 'rematch2', 'mime', 'openssl', 'timechange', 'fansi', 'utf8', 'systemfonts', 'textshaping', 'vroom', 'tzdb', 'progress', 'callr', 'fs', 'knitr', 'rmarkdown', 'selectr', 'stringi', 'broom', 'conflicted', 'cli', 'dbplyr', 'dplyr', 'dtplyr', 'forcats', 'ggplot2', 'googledrive', 'googlesheets4', 'haven', 'hms', 'httr', 'jsonlite', 'lubridate', 'magrittr', 'modelr', 'pillar', 'purrr', 'ragg', 'readr', 'readxl', 'reprex', 'rlang', 'rvest', 'stringr', 'tibble', 'tidyr', 'xml2'
Warning message:
"unable to access index for repository https://cran.r-project.org/bin/windows/contrib/3.6:
cannot open URL 'https://cran.r-project.org/bin/windows/contrib/3.6/PACKAGES'"Packages which are only available in source form, and may need
compilation of C/C++/Fortran: 'fastmap' 'colorspace' 'sys' 'bit' 'ps'
'sass' 'cachem' 'nlme' 'farver' 'rappdirs' 'askpass' 'bit64'
'processx' 'xfun' 'yaml' 'htmltools' 'backports' 'ellipsis' 'glue'
'tidyselect' 'vctrs' 'data.table' 'isoband' 'mgcv' 'scales' 'curl'
'mime' 'openssl' 'timechange' 'fansi' 'utf8' 'systemfonts'
'textshaping' 'vroom' 'tzdb' 'fs' 'stringi' 'cli' 'dplyr' 'haven'
'jsonlite' 'lubridate' 'magrittr' 'purrr' 'ragg' 'readr' 'readxl'
'rlang' 'tibble' 'tidyr' 'xml2'
These will not be installed
installing the source packages 'munsell', 'highr', 'bslib', 'fontawesome', 'tinytex', 'lifecycle', 'memoise', 'blob', 'gtable', 'gargle', 'cellranger', 'ids', 'rematch2', 'progress', 'callr', 'knitr', 'rmarkdown', 'selectr', 'broom', 'conflicted', 'dbplyr', 'dtplyr', 'forcats', 'ggplot2', 'googledrive', 'googlesheets4', 'hms', 'httr', 'modelr', 'pillar', 'reprex', 'rvest', 'stringr', 'tidyverse'
Warning message in install.packages("tidyverse"):
"installation of package 'munsell' had non-zero exit status"Warning message in install.packages("tidyverse"):
"installation of package 'highr' had non-zero exit status"Warning message in install.packages("tidyverse"):
"installation of package 'fontawesome' had non-zero exit status"Warning message in install.packages("tidyverse"):
"installation of package 'tinytex' had non-zero exit status"Warning message in install.packages("tidyverse"):
"installation of package 'lifecycle' had non-zero exit status"Warning message in install.packages("tidyverse"):
"installation of package 'memoise' had non-zero exit status"Warning message in install.packages("tidyverse"):
"installation of package 'blob' had non-zero exit status"Warning message in install.packages("tidyverse"):
"installation of package 'cellranger' had non-zero exit status"Warning message in install.packages("tidyverse"):
"installation of package 'ids' had non-zero exit status"Warning message in install.packages("tidyverse"):
"installation of package 'rematch2' had non-zero exit status"Warning message in install.packages("tidyverse"):
"installation of package 'callr' had non-zero exit status"Warning message in install.packages("tidyverse"):
"installation of package 'httr' had non-zero exit status"Warning message in install.packages("tidyverse"):
"installation of package 'bslib' had non-zero exit status"Warning message in install.packages("tidyverse"):
"installation of package 'gtable' had non-zero exit status"Warning message in install.packages("tidyverse"):
"installation of package 'gargle' had non-zero exit status"Warning message in install.packages("tidyverse"):
"installation of package 'knitr' had non-zero exit status"Warning message in install.packages("tidyverse"):
"installation of package 'conflicted' had non-zero exit status"Warning message in install.packages("tidyverse"):
"installation of package 'dtplyr' had non-zero exit status"Warning message in install.packages("tidyverse"):
"installation of package 'forcats' had non-zero exit status"Warning message in install.packages("tidyverse"):
"installation of package 'hms' had non-zero exit status"Warning message in install.packages("tidyverse"):
"installation of package 'pillar' had non-zero exit status"Warning message in install.packages("tidyverse"):
"installation of package 'stringr' had non-zero exit status"Warning message in install.packages("tidyverse"):
"installation of package 'progress' had non-zero exit status"Warning message in install.packages("tidyverse"):
"installation of package 'rmarkdown' had non-zero exit status"Warning message in install.packages("tidyverse"):
"installation of package 'selectr' had non-zero exit status"Warning message in install.packages("tidyverse"):
"installation of package 'broom' had non-zero exit status"Warning message in install.packages("tidyverse"):
"installation of package 'dbplyr' had non-zero exit status"Warning message in install.packages("tidyverse"):
"installation of package 'ggplot2' had non-zero exit status"Warning message in install.packages("tidyverse"):
"installation of package 'googledrive' had non-zero exit status"Warning message in install.packages("tidyverse"):
"installation of package 'googlesheets4' had non-zero exit status"Warning message in install.packages("tidyverse"):
"installation of package 'modelr' had non-zero exit status"Warning message in install.packages("tidyverse"):
"installation of package 'reprex' had non-zero exit status"Warning message in install.packages("tidyverse"):
"installation of package 'rvest' had non-zero exit status"Warning message in install.packages("tidyverse"):
"installation of package 'tidyverse' had non-zero exit status"Warning message:
"dependencies 'Matrix', 'MASS', 'lattice' are not available"also installing the dependencies 'utf8', 'pillar', 'vctrs', 'glue', 'fs', 'pkgbuild', 'diffobj', 'fansi', 'rematch2', 'tibble', 'brio', 'callr', 'cli', 'desc', 'digest', 'jsonlite', 'lifecycle', 'magrittr', 'pkgload', 'processx', 'ps', 'rlang', 'waldo', 'testthat', 'nlme', 'minqa', 'nloptr', 'RcppEigen'
Warning message:
"unable to access index for repository https://cran.r-project.org/bin/windows/contrib/3.6:
cannot open URL 'https://cran.r-project.org/bin/windows/contrib/3.6/PACKAGES'"Packages which are only available in source form, and may need
compilation of C/C++/Fortran: 'utf8' 'vctrs' 'glue' 'fs' 'diffobj'
'fansi' 'tibble' 'brio' 'cli' 'digest' 'jsonlite' 'magrittr'
'processx' 'ps' 'rlang' 'testthat' 'nlme' 'minqa' 'nloptr'
'RcppEigen' 'lme4'
These will not be installed
installing the source packages 'pillar', 'pkgbuild', 'rematch2', 'callr', 'desc', 'lifecycle', 'pkgload', 'waldo'
Warning message in install.packages("lme4"):
"installation of package 'rematch2' had non-zero exit status"Warning message in install.packages("lme4"):
"installation of package 'callr' had non-zero exit status"Warning message in install.packages("lme4"):
"installation of package 'desc' had non-zero exit status"Warning message in install.packages("lme4"):
"installation of package 'lifecycle' had non-zero exit status"Warning message in install.packages("lme4"):
"installation of package 'pillar' had non-zero exit status"Warning message in install.packages("lme4"):
"installation of package 'pkgbuild' had non-zero exit status"Warning message in install.packages("lme4"):
"installation of package 'waldo' had non-zero exit status"Warning message in install.packages("lme4"):
"installation of package 'pkgload' had non-zero exit status"Warning message:
"unable to access index for repository https://cran.r-project.org/bin/windows/contrib/3.6:
cannot open URL 'https://cran.r-project.org/bin/windows/contrib/3.6/PACKAGES'"installing the source package 'stargazer'
also installing the dependency 'bitops'
Warning message:
"unable to access index for repository https://cran.r-project.org/bin/windows/contrib/3.6:
cannot open URL 'https://cran.r-project.org/bin/windows/contrib/3.6/PACKAGES'"Packages which are only available in source form, and may need
compilation of C/C++/Fortran: 'bitops' 'caTools'
These will not be installed
Warning message:
"dependencies 'lattice', 'MASS', 'Matrix' are not available"also installing the dependencies 'parallelly', 'future', 'future.apply', 'colorspace', 'utf8', 'KernSmooth', 'lava', 'farver', 'munsell', 'fansi', 'pillar', 'tzdb', 'rpart', 'survival', 'nnet', 'prodlim', 'timechange', 'stringi', 'cli', 'glue', 'gtable', 'isoband', 'lifecycle', 'mgcv', 'rlang', 'scales', 'tibble', 'vctrs', 'class', 'proxy', 'data.table', 'dplyr', 'clock', 'ellipsis', 'gower', 'hardhat', 'ipred', 'lubridate', 'magrittr', 'purrr', 'tidyr', 'tidyselect', 'stringr', 'ggplot2', 'e1071', 'ModelMetrics', 'nlme', 'plyr', 'pROC', 'recipes', 'reshape2'
Warning message:
"unable to access index for repository https://cran.r-project.org/bin/windows/contrib/3.6:
cannot open URL 'https://cran.r-project.org/bin/windows/contrib/3.6/PACKAGES'"Packages which are only available in source form, and may need
compilation of C/C++/Fortran: 'parallelly' 'colorspace' 'utf8'
'KernSmooth' 'farver' 'fansi' 'tzdb' 'rpart' 'survival' 'nnet'
'prodlim' 'timechange' 'stringi' 'cli' 'glue' 'isoband' 'mgcv'
'rlang' 'scales' 'tibble' 'vctrs' 'class' 'proxy' 'data.table'
'dplyr' 'clock' 'ellipsis' 'gower' 'ipred' 'lubridate' 'magrittr'
'purrr' 'tidyr' 'tidyselect' 'e1071' 'ModelMetrics' 'nlme' 'plyr'
'pROC' 'reshape2' 'caret'
These will not be installed
installing the source packages 'future', 'future.apply', 'lava', 'munsell', 'pillar', 'gtable', 'lifecycle', 'hardhat', 'stringr', 'ggplot2', 'recipes'
Warning message in install.packages("caret"):
"installation of package 'future' had non-zero exit status"Warning message in install.packages("caret"):
"installation of package 'munsell' had non-zero exit status"Warning message in install.packages("caret"):
"installation of package 'lifecycle' had non-zero exit status"Warning message in install.packages("caret"):
"installation of package 'hardhat' had non-zero exit status"Warning message in install.packages("caret"):
"installation of package 'future.apply' had non-zero exit status"Warning message in install.packages("caret"):
"installation of package 'pillar' had non-zero exit status"Warning message in install.packages("caret"):
"installation of package 'gtable' had non-zero exit status"Warning message in install.packages("caret"):
"installation of package 'stringr' had non-zero exit status"Warning message in install.packages("caret"):
"installation of package 'recipes' had non-zero exit status"Warning message in install.packages("caret"):
"installation of package 'lava' had non-zero exit status"Warning message in install.packages("caret"):
"installation of package 'ggplot2' had non-zero exit status"Warning message:
"dependency 'Matrix' is not available"also installing the dependencies 'survival', 'RcppEigen'
Warning message:
"unable to access index for repository https://cran.r-project.org/bin/windows/contrib/3.6:
cannot open URL 'https://cran.r-project.org/bin/windows/contrib/3.6/PACKAGES'"Packages which are only available in source form, and may need
compilation of C/C++/Fortran: 'survival' 'RcppEigen' 'glmnet'
These will not be installed
library(tidyverse)
library(lme4)
library(stargazer)
library(caTools)
library(caret)
library(glmnet)
library(rsample)
library(hdm)
Error in library(tidyverse): there is no package called 'tidyverse'
Traceback:
1. library(tidyverse)
df <- read.csv("./data/bruhn2016.csv")
first(df, 5)
| Row | outcome.test.score | treatment | school | is.female | mother.attended.secondary.school | father.attened.secondary.school | failed.at.least.one.school.year | family.receives.cash.transfer | has.computer.with.internet.at.home | is.unemployed | has.some.form.of.income | saves.money.for.future.purchases | intention.to.save.index | makes.list.of.expenses.every.month | negotiates.prices.or.payment.methods | financial.autonomy.index |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Float64 | Int64 | Int64 | String3 | String3 | String3 | String3 | String3 | String3 | String3 | String3 | String3 | String3 | String3 | String3 | String3 | |
| 1 | 47.3674 | 0 | 17018390 | NA | NA | NA | NA | NA | NA | 1 | 1 | 0 | 29 | 0 | 1 | 52 |
| 2 | 58.1768 | 1 | 33002614 | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 41 | 0 | 0 | 27 |
| 3 | 56.6717 | 1 | 35002914 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 48 | 0 | 1 | 56 |
| 4 | 29.0794 | 0 | 35908915 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 42 | 0 | 0 | 27 |
| 5 | 49.5635 | 1 | 33047324 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 50 | 0 | 1 | 31 |
# Drop missing values
df <- df %>% drop_na()
df
Error in library(dplyr): there is no package called 'dplyr'
Traceback:
1. library(dplyr)
missing_count = sum(ismissing, eachcol(df))
0
# Define the vector of dependent variable names
dependent_vars <- c("outcome.test.score", "intention.to.save.index", "negotiates.prices.or.payment.methods", "has.some.form.of.income", "makes.list.of.expenses.every.month", "financial.autonomy.index", "saves.money.for.future.purchases", "is.unemployed")
["outcome.test.score", "treatment", "school", "is.female", "mother.attended.secondary.school", "father.attened.secondary.school", "failed.at.least.one.school.year", "family.receives.cash.transfer", "has.computer.with.internet.at.home", "is.unemployed", "has.some.form.of.income", "saves.money.for.future.purchases", "intention.to.save.index", "makes.list.of.expenses.every.month", "negotiates.prices.or.payment.methods", "financial.autonomy.index"]
# Define an array of dependent variable names
ddependent_vars <- c("outcome.test.score",
"intention.to.save.index",
"negotiates.prices.or.payment.methods",
"has.some.form.of.income",
"makes.list.of.expenses.every.month",
"financial.autonomy.index",
"saves.money.for.future.purchases",
"is.unemployed")
8-element Vector{String}:
"outcome.test.score"
"intention.to.save.index"
"negotiates.prices.or.payment.methods"
"has.some.form.of.income"
"makes.list.of.expenses.every.month"
"financial.autonomy.index"
"saves.money.for.future.purchases"
"is.unemployed"
For Lasso regressions, we split the data into train and test data, and standarize the covariates matrix
X <- df[, !(names(df) %in% dependent_vars)]
y <- df[, (names(df) %in% dependent_vars)]
set.seed(42)
# Split the data into training and testing sets
trainIndex <- createDataPartition(y[,1], p = 0.8, list = FALSE)
X_train <- X[trainIndex, ]
X_test <- X[-trainIndex, ]
y_train <- y[trainIndex, ]
y_test <- y[-trainIndex, ]
# Extract 'treatment' and remove it from 'X_train' and 'X_test'
T_train <- X_train$treatment
T_test <- X_test$treatment
X_train <- X_train[, !names(X_train) %in% "treatment"]
X_test <- X_test[, !names(X_test) %in% "treatment"]
# Standardize the covariates matrix
scale <- preProcess(X_train, method = c("center", "scale"))
X_train_scaled <- predict(scale, X_train)
X_test_scaled <- predict(scale, X_test)
# Combine the scaled training and testing data
X_scaled <- rbind(X_train_scaled, X_test_scaled) %>%
arrange(rownames(.))
# Combine the treatment training and testing data
T <- rbind(T_train, T_test) %>%
arrange(rownames(.))
2.2.2. Regressions#
a. OLS#
From 1 - 3 regression: measures treatment impact on student financial proficiency
From 4 - 6 regression: measures treatment impact on student savings behavior and attitudes
From 7 - 9 regression: measures treatment impact on student money management behavior and attitudes
From 10 - 12 regression: measures treatment impact on student entrepreneurship and work outcomes
# OLS regressions with "Student Financial Proficiency" as dependent variable
ols_score_1 <- lm(outcome.test.score ~ treatment, data = df)
ols_score_2 <- lm(outcome.test.score ~ treatment + school + failed.at.least.one.school.year, data = df)
ols_score_3 <- lm(outcome.test.score ~ treatment + school + failed.at.least.one.school.year + is.female + mother.attended.secondary.school + father.attended.secondary.school + family.receives.cash.transfer + has.computer.with.internet.at.home, data = df)
# OLS regressions with "Intention to save index" as dependent variable
ols_saving_1 <- lm(intention.to.save.index ~ treatment, data = df)
ols_saving_2 <- lm(intention.to.save.index ~ treatment + school + failed.at.least.one.school.year, data = df)
ols_saving_3 <- lm(intention.to.save.index ~ treatment + school + failed.at.least.one.school.year + is.female + mother.attended.secondary.school + father.attended.secondary.school + family.receives.cash.transfer + has.computer.with.internet.at.home, data = df)
# OLS regressions with "Negotiates prices or payment methods" as dependent variable
ols_negotiates_1 <- lm(negotiates.prices.or.payment.methods ~ treatment, data = df)
ols_negotiates_2 <- lm(negotiates.prices.or.payment.methods ~ treatment + school + failed.at.least.one.school.year, data = df)
ols_negotiates_3 <- lm(negotiates.prices.or.payment.methods ~ treatment + school + failed.at.least.one.school.year + is.female + mother.attended.secondary.school + father.attended.secondary.school + family.receives.cash.transfer + has.computer.with.internet.at.home, data = df)
# OLS regressions with "Has some form of income" as dependent variable
ols_manage_1 <- lm(has.some.form.of.income ~ treatment, data = df)
ols_manage_2 <- lm(has.some.form.of.income ~ treatment + school + failed.at.least.one.school.year, data = df)
ols_manage_3 <- lm(has.some.form.of.income ~ treatment + school + failed.at.least.one.school.year + is.female + mother.attended.secondary.school + father.attended.secondary.school + family.receives.cash.transfer + has.computer.with.internet.at.home, data = df)
# Show parameters in table using stargazer
stargazer(ols_score_1, ols_score_2, ols_score_3, ols_saving_1, ols_saving_2, ols_saving_3, ols_negotiates_1, ols_negotiates_2, ols_negotiates_3, ols_manage_1, ols_manage_2, ols_manage_3, type = "text",
title = "Regression Results",
column.labels = c("Student Financial Proficiency", "Intention to save index", "Negotiates prices or payment methods", "Has some form of income"),
covariate.labels = c("Failed at least one school year", "Female", "Father attended secondary school", "Family receives cash transfer", "Has computer with internet at home"))
Intel MKL WARNING: Support of Intel(R) Streaming SIMD Extensions 4.2 (Intel(R) SSE4.2) enabled only processors has been deprecated. Intel oneAPI Math Kernel Library 2025.0 will require Intel(R) Advanced Vector Extensions (Intel(R) AVX) instructions.
Intel MKL WARNING: Support of Intel(R) Streaming SIMD Extensions 4.2 (Intel(R) SSE4.2) enabled only processors has been deprecated. Intel oneAPI Math Kernel Library 2025.0 will require Intel(R) Advanced Vector Extensions (Intel(R) AVX) instructions.
Intel MKL WARNING: Support of Intel(R) Streaming SIMD Extensions 4.2 (Intel(R) SSE4.2) enabled only processors has been deprecated. Intel oneAPI Math Kernel Library 2025.0 will require Intel(R) Advanced Vector Extensions (Intel(R) AVX) instructions.
Intel MKL WARNING: Support of Intel(R) Streaming SIMD Extensions 4.2 (Intel(R) SSE4.2) enabled only processors has been deprecated. Intel oneAPI Math Kernel Library 2025.0 will require Intel(R) Advanced Vector Extensions (Intel(R) AVX) instructions.
Intel MKL WARNING: Support of Intel(R) Streaming SIMD Extensions 4.2 (Intel(R) SSE4.2) enabled only processors has been deprecated. Intel oneAPI Math Kernel Library 2025.0 will require Intel(R) Advanced Vector Extensions (Intel(R) AVX) instructions.
Intel MKL WARNING: Support of Intel(R) Streaming SIMD Extensions 4.2 (Intel(R) SSE4.2) enabled only processors has been deprecated. Intel oneAPI Math Kernel Library 2025.0 will require Intel(R) Advanced Vector Extensions (Intel(R) AVX) instructions.
Intel MKL WARNING: Support of Intel(R) Streaming SIMD Extensions 4.2 (Intel(R) SSE4.2) enabled only processors has been deprecated. Intel oneAPI Math Kernel Library 2025.0 will require Intel(R) Advanced Vector Extensions (Intel(R) AVX) instructions.
Intel MKL WARNING: Support of Intel(R) Streaming SIMD Extensions 4.2 (Intel(R) SSE4.2) enabled only processors has been deprecated. Intel oneAPI Math Kernel Library 2025.0 will require Intel(R) Advanced Vector Extensions (Intel(R) AVX) instructions.
Intel MKL WARNING: Support of Intel(R) Streaming SIMD Extensions 4.2 (Intel(R) SSE4.2) enabled only processors has been deprecated. Intel oneAPI Math Kernel Library 2025.0 will require Intel(R) Advanced Vector Extensions (Intel(R) AVX) instructions.
Intel MKL WARNING: Support of Intel(R) Streaming SIMD Extensions 4.2 (Intel(R) SSE4.2) enabled only processors has been deprecated. Intel oneAPI Math Kernel Library 2025.0 will require Intel(R) Advanced Vector Extensions (Intel(R) AVX) instructions.
Intel MKL WARNING: Support of Intel(R) Streaming SIMD Extensions 4.2 (Intel(R) SSE4.2) enabled only processors has been deprecated. Intel oneAPI Math Kernel Library 2025.0 will require Intel(R) Advanced Vector Extensions (Intel(R) AVX) instructions.
Intel MKL WARNING: Support of Intel(R) Streaming SIMD Extensions 4.2 (Intel(R) SSE4.2) enabled only processors has been deprecated. Intel oneAPI Math Kernel Library 2025.0 will require Intel(R) Advanced Vector Extensions (Intel(R) AVX) instructions.
| Dependent var 1: Student Financial Proficiency | Dependent var 2: Intention to save index | Dependent var 3: Negotiates prices or payment methods | Dependent var 4: Has some form of income | |||||||||
| (1) | (2) | (3) | (4) | (5) | (6) | (7) | (8) | (9) | (10) | (11) | (12) | |
| Intercept | 57.591*** | 59.377*** | 57.013*** | 49.016*** | 46.725*** | 45.407*** | 0.763*** | 0.856*** | 0.901*** | 0.639*** | 0.534*** | 0.553*** |
| (0.187) | (0.556) | (0.583) | (0.240) | (0.728) | (0.767) | (0.006) | (0.017) | (0.018) | (0.006) | (0.019) | (0.020) | |
| Failed at least one school year | -7.218*** | -6.759*** | -3.614*** | -3.354*** | 0.024*** | 0.016* | 0.005 | 0.002 | ||||
| (0.288) | (0.289) | (0.377) | (0.380) | (0.009) | (0.009) | (0.010) | (0.010) | |||||
| Female | 2.836*** | 1.357*** | -0.066*** | -0.054*** | ||||||||
| (0.257) | (0.339) | (0.008) | (0.009) | |||||||||
| Q("mother.attended.secondary.school") | 1.826*** | 1.380*** | -0.016** | 0.036*** | ||||||||
| (0.257) | (0.338) | (0.008) | (0.009) | |||||||||
| school | 0.000 | -0.000 | 0.000*** | 0.000*** | -0.000*** | -0.000*** | 0.000*** | 0.000*** | ||||
| (0.000) | (0.000) | (0.000) | (0.000) | (0.000) | (0.000) | (0.000) | (0.000) | |||||
| treatment | 4.216*** | 4.392*** | 4.307*** | -0.070 | -0.005 | -0.047 | 0.001 | 0.001 | 0.003 | 0.017** | 0.016* | 0.018** |
| (0.261) | (0.255) | (0.253) | (0.335) | (0.334) | (0.333) | (0.008) | (0.008) | (0.008) | (0.009) | (0.009) | (0.009) | |
| Observations | 12222 | 12222 | 12222 | 12222 | 12222 | 12222 | 12222 | 12222 | 12222 | 12222 | 12222 | 12222 |
| R2 | 0.021 | 0.069 | 0.081 | 0.000 | 0.009 | 0.012 | 0.000 | 0.004 | 0.010 | 0.000 | 0.003 | 0.008 |
| Adjusted R2 | 0.021 | 0.068 | 0.080 | -0.000 | 0.009 | 0.011 | -0.000 | 0.004 | 0.010 | 0.000 | 0.003 | 0.008 |
| Residual Std. Error | 14.432 (df=12220) | 14.076 (df=12218) | 13.986 (df=12216) | 18.506 (df=12220) | 18.421 (df=12218) | 18.400 (df=12216) | 0.425 (df=12220) | 0.424 (df=12218) | 0.423 (df=12216) | 0.478 (df=12220) | 0.477 (df=12218) | 0.476 (df=12216) |
| F Statistic | 260.547*** (df=1; 12220) | 300.463*** (df=3; 12218) | 214.786*** (df=5; 12216) | 0.043 (df=1; 12220) | 38.533*** (df=3; 12218) | 29.248*** (df=5; 12216) | 0.018 (df=1; 12220) | 16.352*** (df=3; 12218) | 24.653*** (df=5; 12216) | 3.843** (df=1; 12220) | 12.839*** (df=3; 12218) | 19.652*** (df=5; 12216) |
| Note: | *p<0.1; **p<0.05; ***p<0.01 | |||||||||||
b. Double Lasso using cross validation#
Dependent var 1: Student Financial Proficiency
Step 1: We ran Lasso regression of Y (student financial proficiency) on X, and T on X
#we need matix structure
X_matrix <- as.matrix(X_scaled)
y_vector <- y_train$outcome.test.score
T_vector <- as.numeric(T_train)
# Lasso regression of Y on X with cross-validation
lasso_CV_yX <- cv.glmnet(X_matrix, y_vector, alpha = 1, lambda = seq(0.0001, 0.5, by = 0.001), nfolds = 10, maxit = 5000)
best_lambda_yX <- lasso_CV_yX$lambda.min
print(sprintf("Best lambda for Y on X: %.4f", best_lambda_yX))
# Estimate Y predictions with all X
y_pred_yX <- predict(lasso_CV_yX, s = best_lambda_yX, newx = as.matrix(X_scaled))
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[4], line 2
1 # Estimate y predictions with all X
----> 2 y_pred_yX = lasso_CV_yX.predict(X_scaled)
NameError: name 'lasso_CV_yX' is not defined
lasso_CV_TX <- cv.glmnet(X_matrix, T_vector, alpha = 1, lambda = seq(0.0001, 0.5, by = 0.001), nfolds = 10, maxit = 5000)
best_lambda_TX <- lasso_CV_TX$lambda.min
print(sprintf("Best lambda for T on X: %.4f", best_lambda_TX))
Intel MKL WARNING: Support of Intel(R) Streaming SIMD Extensions 4.2 (Intel(R) SSE4.2) enabled only processors has been deprecated. Intel oneAPI Math Kernel Library 2025.0 will require Intel(R) Advanced Vector Extensions (Intel(R) AVX) instructions.
Intel MKL WARNING: Support of Intel(R) Streaming SIMD Extensions 4.2 (Intel(R) SSE4.2) enabled only processors has been deprecated. Intel oneAPI Math Kernel Library 2025.0 will require Intel(R) Advanced Vector Extensions (Intel(R) AVX) instructions.
Intel MKL WARNING: Support of Intel(R) Streaming SIMD Extensions 4.2 (Intel(R) SSE4.2) enabled only processors has been deprecated. Intel oneAPI Math Kernel Library 2025.0 will require Intel(R) Advanced Vector Extensions (Intel(R) AVX) instructions.
Intel MKL WARNING: Support of Intel(R) Streaming SIMD Extensions 4.2 (Intel(R) SSE4.2) enabled only processors has been deprecated. Intel oneAPI Math Kernel Library 2025.0 will require Intel(R) Advanced Vector Extensions (Intel(R) AVX) instructions.
Intel MKL WARNING: Support of Intel(R) Streaming SIMD Extensions 4.2 (Intel(R) SSE4.2) enabled only processors has been deprecated. Intel oneAPI Math Kernel Library 2025.0 will require Intel(R) Advanced Vector Extensions (Intel(R) AVX) instructions.
Intel MKL WARNING: Support of Intel(R) Streaming SIMD Extensions 4.2 (Intel(R) SSE4.2) enabled only processors has been deprecated. Intel oneAPI Math Kernel Library 2025.0 will require Intel(R) Advanced Vector Extensions (Intel(R) AVX) instructions.
Intel MKL WARNING: Support of Intel(R) Streaming SIMD Extensions 4.2 (Intel(R) SSE4.2) enabled only processors has been deprecated. Intel oneAPI Math Kernel Library 2025.0 will require Intel(R) Advanced Vector Extensions (Intel(R) AVX) instructions.
Intel MKL WARNING: Support of Intel(R) Streaming SIMD Extensions 4.2 (Intel(R) SSE4.2) enabled only processors has been deprecated. Intel oneAPI Math Kernel Library 2025.0 will require Intel(R) Advanced Vector Extensions (Intel(R) AVX) instructions.
Intel MKL WARNING: Support of Intel(R) Streaming SIMD Extensions 4.2 (Intel(R) SSE4.2) enabled only processors has been deprecated. Intel oneAPI Math Kernel Library 2025.0 will require Intel(R) Advanced Vector Extensions (Intel(R) AVX) instructions.
Intel MKL WARNING: Support of Intel(R) Streaming SIMD Extensions 4.2 (Intel(R) SSE4.2) enabled only processors has been deprecated. Intel oneAPI Math Kernel Library 2025.0 will require Intel(R) Advanced Vector Extensions (Intel(R) AVX) instructions.
Mejor lambda: 0.0011
# Estimate T predictions with all X
y_pred_TX <- predict(lasso_CV_TX, s = best_lambda_TX, newx = as.matrix(X_scaled))
Step 2: Obtain the resulting residuals
#y_pred_yX <- as.numeric(y_pred_yX)
#y_pred_TX <- as.numeric(y_pred_TX)
res_yX <- y$outcome.test.score - y_pred_yX
res_TX <- T - y_pred_TX
Step 3: We run the least squares of res_yX on res_TX
# Create a data frame for the residuals
residuals_data <- data.frame(res_yX = res_yX, res_TX = res_TX)
ols_score <- lm(res_yX ~ res_TX, data = residuals_data)
stargazer(ols_score, type = "text", title = "OLS Regression of Residuals")
Intel MKL WARNING: Support of Intel(R) Streaming SIMD Extensions 4.2 (Intel(R) SSE4.2) enabled only processors has been deprecated. Intel oneAPI Math Kernel Library 2025.0 will require Intel(R) Advanced Vector Extensions (Intel(R) AVX) instructions.
| Dependent variable: res_yX | |
| (1) | |
| Intercept | 0.033 |
| (0.126) | |
| res_TX | 4.324*** |
| (0.253) | |
| Observations | 12222 |
| R2 | 0.023 |
| Adjusted R2 | 0.023 |
| Residual Std. Error | 13.945 (df=12220) |
| F Statistic | 292.956*** (df=1; 12220) |
| Note: | *p<0.1; **p<0.05; ***p<0.01 |
c. Double Lasso using theoretical lambda#
# Load necessary libraries
library(hdm)
# Define the RLasso class
RLasso <- setRefClass("RLasso",
fields = list(post = "logical"),
methods = list(
initialize = function(post = TRUE) {
.self$post <- post
},
fit = function(X, y) {
fit <- rlasso(X, y, post = .self$post)
.self$rlasso_ <- fit
return(.self)
},
predict = function(X) {
predictions <- predict(.self$rlasso_, newx = X)
return(as.matrix(X) %*% predictions$coefficients[-1] + predictions$coefficients[1])
},
nsel = function() {
sum(abs(coef(.self$rlasso_)[-1]) > 0)
}
)
)
lasso_model <- function() {
RLasso$new(post = FALSE)
}
model <- lasso_model()
model$fit(X, y)
y_pred <- model$predict(X)
Step 1:
# Assuming X_scaled, y, and T are prepared as in Python
# Estimate y predictions with all X
model_yX <- lasso_model()
model_yX$fit(X_scaled, y$outcome.test.score)
y_pred_yX <- model_yX$predict(X_scaled)
Intel MKL WARNING: Support of Intel(R) Streaming SIMD Extensions 4.2 (Intel(R) SSE4.2) enabled only processors has been deprecated. Intel oneAPI Math Kernel Library 2025.0 will require Intel(R) Advanced Vector Extensions (Intel(R) AVX) instructions.
Intel MKL WARNING: Support of Intel(R) Streaming SIMD Extensions 4.2 (Intel(R) SSE4.2) enabled only processors has been deprecated. Intel oneAPI Math Kernel Library 2025.0 will require Intel(R) Advanced Vector Extensions (Intel(R) AVX) instructions.
Intel MKL WARNING: Support of Intel(R) Streaming SIMD Extensions 4.2 (Intel(R) SSE4.2) enabled only processors has been deprecated. Intel oneAPI Math Kernel Library 2025.0 will require Intel(R) Advanced Vector Extensions (Intel(R) AVX) instructions.
Intel MKL WARNING: Support of Intel(R) Streaming SIMD Extensions 4.2 (Intel(R) SSE4.2) enabled only processors has been deprecated. Intel oneAPI Math Kernel Library 2025.0 will require Intel(R) Advanced Vector Extensions (Intel(R) AVX) instructions.
# Estimate T predictions with all X
model_TX <- lasso_model()
model_TX$fit(X_scaled, T)
y_pred_TX <- model_TX$predict(X_scaled)
Intel MKL WARNING: Support of Intel(R) Streaming SIMD Extensions 4.2 (Intel(R) SSE4.2) enabled only processors has been deprecated. Intel oneAPI Math Kernel Library 2025.0 will require Intel(R) Advanced Vector Extensions (Intel(R) AVX) instructions.
Intel MKL WARNING: Support of Intel(R) Streaming SIMD Extensions 4.2 (Intel(R) SSE4.2) enabled only processors has been deprecated. Intel oneAPI Math Kernel Library 2025.0 will require Intel(R) Advanced Vector Extensions (Intel(R) AVX) instructions.
Intel MKL WARNING: Support of Intel(R) Streaming SIMD Extensions 4.2 (Intel(R) SSE4.2) enabled only processors has been deprecated. Intel oneAPI Math Kernel Library 2025.0 will require Intel(R) Advanced Vector Extensions (Intel(R) AVX) instructions.
Intel MKL WARNING: Support of Intel(R) Streaming SIMD Extensions 4.2 (Intel(R) SSE4.2) enabled only processors has been deprecated. Intel oneAPI Math Kernel Library 2025.0 will require Intel(R) Advanced Vector Extensions (Intel(R) AVX) instructions.
Step 2:
res_yX <- y$outcome.test.score - y_pred_yX
res_TX <- T - y_pred_TX
Step 3:
library(lmtest)
library(sandwich)
library(stargazer)
df <- data.frame(res_yX, res_TX)
lasso_hdm_score <- lm(res_yX ~ res_TX, data = df)
summary(lasso_hdm_score)
# Display the regression table using stargazer
stargazer(lasso_hdm_score, type = "text")
Intel MKL WARNING: Support of Intel(R) Streaming SIMD Extensions 4.2 (Intel(R) SSE4.2) enabled only processors has been deprecated. Intel oneAPI Math Kernel Library 2025.0 will require Intel(R) Advanced Vector Extensions (Intel(R) AVX) instructions.
| Dependent variable: res_yX | |
| (1) | |
| Intercept | 0.000 |
| (0.126) | |
| res_TX | 4.316*** |
| (0.253) | |
| Observations | 12222 |
| R2 | 0.023 |
| Adjusted R2 | 0.023 |
| Residual Std. Error | 13.953 (df=12220) |
| F Statistic | 291.837*** (df=1; 12220) |
| Note: | *p<0.1; **p<0.05; ***p<0.01 |
# Save the ITT beta and the confidence intervals
beta_DL_theo <- coef(lasso_hdm_score)['res_TX']
conf_int_DL_theo <- confint(lasso_hdm_score, 'res_TX')
d. Double Lasso using partialling out method#
rlassoEffect = hdmpy.rlassoEffect(X_scaled, y['outcome.test.score'], T, method='partialling out')
rlassoEffect
beta_part_out = rlassoEffect['coefficient']
critical_value = 1.96 # For 95% confidence level
conf_int_part_out = [beta_part_out - critical_value * rlassoEffect['se'],
beta_part_out + critical_value * rlassoEffect['se']]
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[1], line 1
----> 1 rlassoEffect = hdmpy.rlassoEffect(X_scaled, y['outcome.test.score'], T, method='partialling out')
NameError: name 'hdmpy' is not defined
rlassoEffect <- rlassoEffect(x = X_scaled, y = y$outcome.test.score, d = T, method = "partiallingOut")
{'alpha': 4.313441,
'se': array([0.25271166]),
't': array([17.06862565]),
'pval': array([2.54111627e-65]),
'coefficients': 4.313441,
'coefficient': 4.313441,
'coefficients_reg': 0
(Intercept) 59.769260
x0 0.000000
x1 1.511205
x2 0.529423
x3 0.461196
x4 -2.878027
x5 -0.857569
x6 0.000000,
'selection_index': array([[False],
[ True],
[ True],
[ True],
[ True],
[ True],
[False]]),
'residuals': {'epsilon': array([[-10.04200277],
[-31.30841071],
[-15.13769394],
...,
[-17.22383794],
[ -3.93047339],
[ -4.88461742]]),
'v': array([[ 0.48682705],
[-0.513173 ],
[ 0.48682705],
...,
[ 0.48682705],
[-0.513173 ],
[-0.513173 ]], dtype=float32)},
'samplesize': 12222}
print(rlassoEffect)
critical_value <- 1.96
conf_int_part_out <- c(beta_part_out - critical_value * summary(rlassoEffect)$se,
beta_part_out + critical_value * summary(rlassoEffect)$se)
print(conf_int_part_out)
Results#
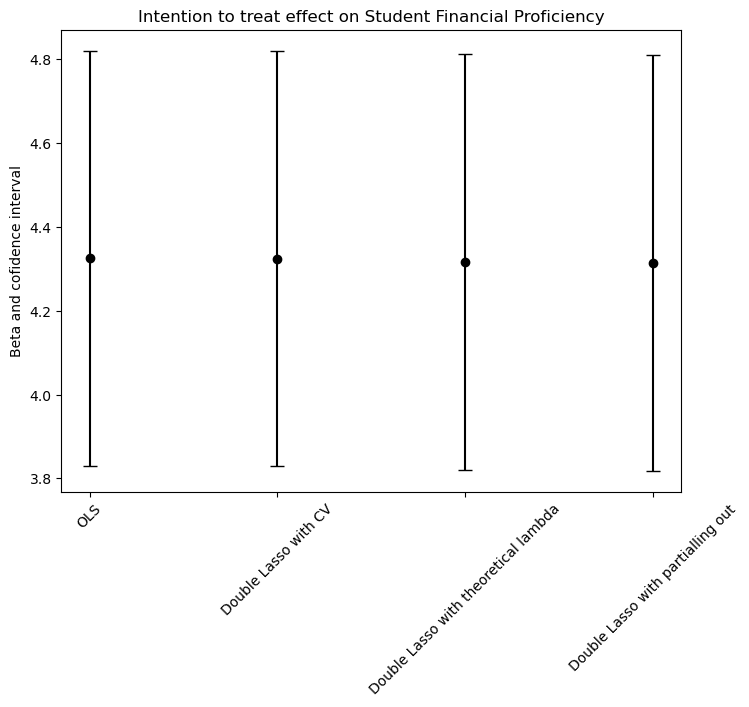
We found that the intention to treat effect (ITT) is very similar estimating with all 4 models (aproximately 4.3, with 95% of confidence). This could be because the ratio between the parameters and the number of observations p/n is small (8/12222 = 0.00065455735). In other words, we are not dealing with high dimensional data and the models from b. to d. will outperform the OLS when we are in the opposite scenario. In conclusion, we can say that the OLS model estimates the ITT just as good as the other models.
library(ggplot2)
data <- data.frame(
Model = c("OLS", "Double Lasso with CV", "Double Lasso with theoretical lambda", "Double Lasso with partialling out"),
Estimate = c(beta_OLS, beta_DL_CV, beta_DL_theo, beta_part_out),
Lower_CI = c(conf_int_OLS[1], conf_int_DL_CV[1], conf_int_DL_theo[1], conf_int_part_out[1]),
Upper_CI = c(conf_int_OLS[2], conf_int_DL_CV[2], conf_int_DL_theo[2], conf_int_part_out[2])
)
# Calculate the error sizes as required for geom_errorbar
data$ymin <- data$Estimate - data$Lower_CI
data$ymax <- data$Upper_CI - data$Estimate
# Create the plot
ggplot(data, aes(x = Model, y = Estimate, ymin = ymin, ymax = ymax)) +
geom_point() +
geom_errorbar(width = 0.2, size = 1, color = "black", position = position_dodge(0.05)) +
labs(title = "Intention to Treat Effect on Student Financial Proficiency",
y = "Beta and Confidence Interval") +
theme_minimal() +
theme(axis.text.x = element_text(angle = 45, hjust = 1))
ggsave("ITT_Effect_Size_Plot.png", width = 8, height = 6, units = "in")